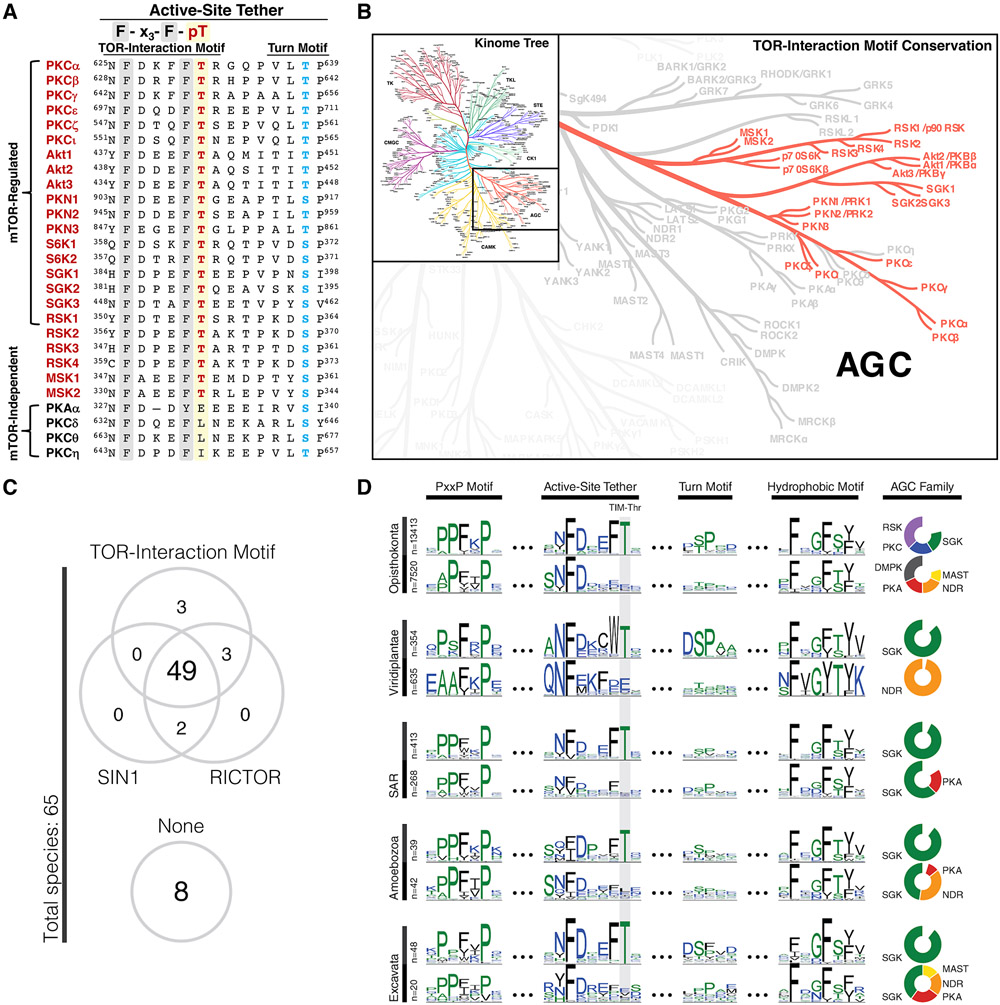
Fig. 4. The TOR-Interaction Motif Is Evolutionarily Conserved in Eukaryotes.
(A) Sequence alignment of the active-site tether region of AGC kinases indicating TOR-interaction motif (red) and turn motif (blue) phosphorylation sites for selected AGC kinases.
(B) AGC kinase branch of the human Kinome tree (111) indicating conservation of the TOR-interaction motif Thr in the highlighted kinases.
(C) Co-conservation of the TOR-interaction motif with TORC2 components SIN1 and RICTOR in various species (detailed in Fig. S7). Conservation of the TOR-interaction motif was defined by species that showed at least one AGC kinase conserving the TIM Thr.
(D) Conservation of the TOR-interaction motif Thr (TIM-Thr) in the AGC C-terminal tail across 5 major taxonomic groups. Sequence logos are shown for four distinct regions of the AGC tail: PxxP motif (124), active-site tether (57), turn motif, and hydrophobic motif. Sequences are stratified by taxonomic clades across the Y-axis: opisthokonta (animals, fungi, yeast), viridiplantae (terrestrial & aquatic plants), SAR (protozoa), amoebozoa (protozoa), and excavata (protozoa). Within each clade, sequences are further stratified by conservation of the TIM-Thr (top row). Pie charts (right) show the composition of AGC families within strata. AGC families accounting for less than 10% of each strata are not shown.