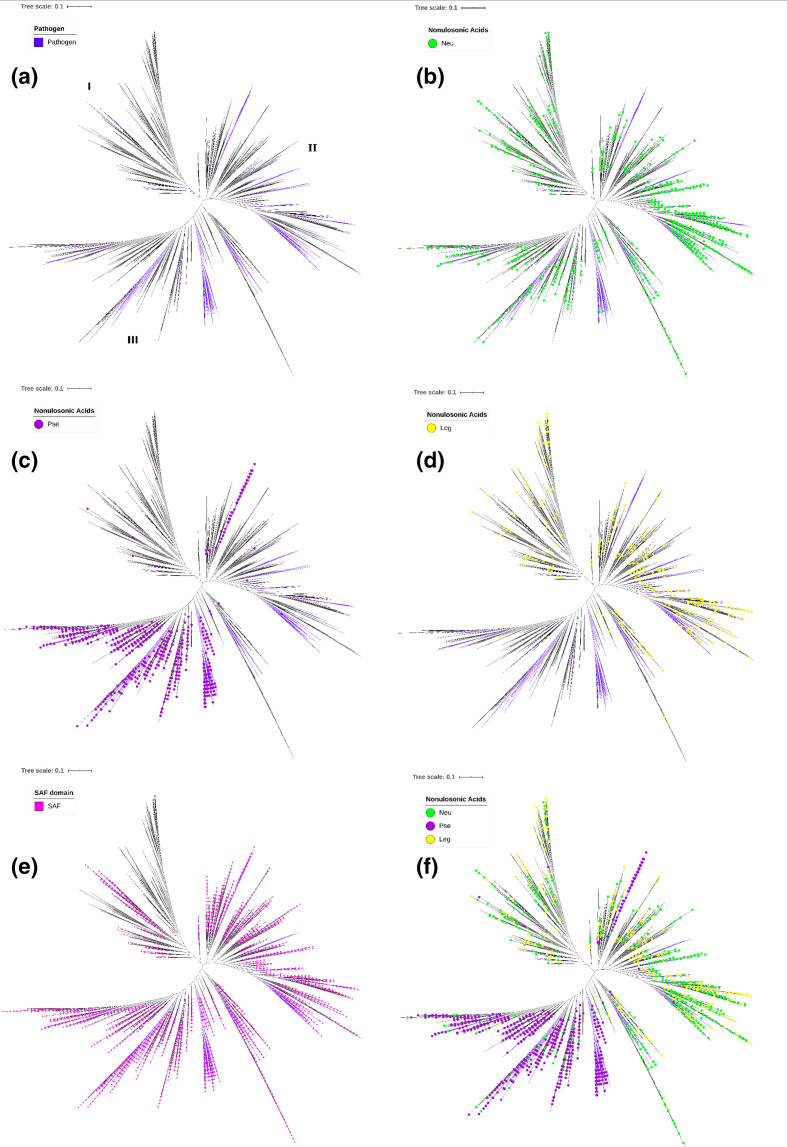
Fig. 4.
Phylogenetic analysis of bacterial NeuB and NeuB homologues. The NeuB HMM profile, reconstructed with the sequences from the KEGG database, was used to mine NeuB sequences and their homologues in the UniprotKB database. A total of 13 941 sequences were recovered and the presence of the NeuB domain was confirmed by comparison with the CDD database. The NeuB domains (~250 aa) were separated from the rest of the sequences and used in phylogenetic analysis. The total neighbour-joining tree was calculated using SWeeP [38], an alignment-free clustering tool. (a) The coloured clades represent the pathogenic organisms identified through the GOLD and KEGG databases. NeuB sequences and homologues were identified based on an unambiguous E.C. number. The coloured dots represent sequences identified as: (b) neuraminic acid synthase (Neu), (c) pseudaminic acid synthase (Pse) and (d) legionaminic acid synthase (Leg). (e) SAF domain identified by Interpro. (f) All previous E.C. numbers identified. Bars, 0.1 amino acid substitutions per site per million years.