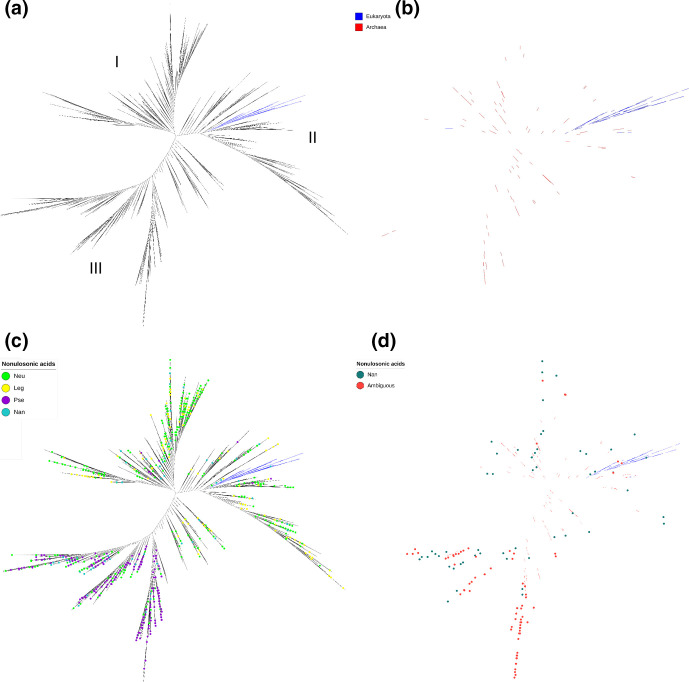
Fig. 5.
Phylogenetic analysis of NeuB, NanS and NeuB homologues. We used the NeuB and NanS sequences retrieved from the KEGG database to create two individual HMM profiles, one for NeuB and the other for NanS. These profiles were used to mine the UniProt database and retrieve more NeuB and NanS sequences. A chi-square distribution was performed in the retrieved data using IQ-Tree, resulting in 7714 sequences. The presence of the NeuB domain was confirmed by comparison with the CDD. The sequences were aligned using MAFFT and an ML tree was inferred using the RaXML tool with 1000 fast bootstrap replicates. (a) ML tree of prokaryotic NeuB and eukaryotic NanS inferred using RaXML. The groups (i) predominantly environmental, (II) intermediate and (III) predominantly pathogenic are marked on the tree. (b) Branches representing NeuB from Archaea and NanS from Eukarya. The blue branches represent Eukarya sequences, the red branches represent Archaea sequences. (c) Phylogenetic tree of NeuB and NanS highlighting sequences with unambiguous E.C. number. (d) Phylogenetic tree of NeuB and NanS highlighting sequences with ambiguous E.C. numbers (multiple identifications for one sequence).