Fig. 1. Response to IL-4 is highly divergent in bone marrow–derived macrophages from different mouse strains.
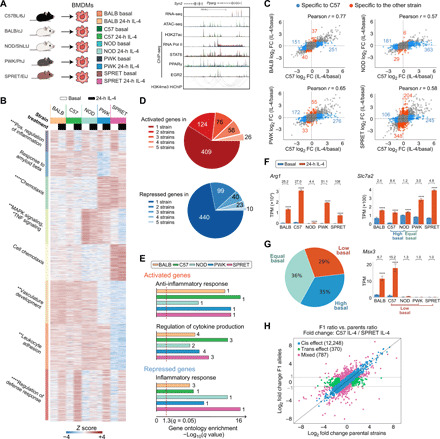
(A) Overview of experimental design and main datasets. (B) WGCNA clustering focused on strain-differentially regulated genes in IL-4–treated bone marrow–derived macrophages (BMDMs). The top hit Metascape pathways are annotated for each module. *q < 0.05, **q < 0.01, ***q < 0.001, ****q < 0.0001. MAPK, mitogen-activated protein kinase; TNF, tumor necrosis factor. (C) Ratio-ratio plots demonstrating the mRNA response to IL-4 in pairwise comparisons. (D) Overlap of genes significantly induced or repressed (q < 0.05, >2-fold) after IL-4 treatment in BMDMs from all strains. (E) Gene ontology terms enriched in up- and down-regulated genes after 24-hour IL-4 stimulation in BMDMs from all strains. Numbers indicate the rank order in pathway analysis. (F) Arg1, Slc7a2, and Msx3 as example genes differentially up-regulated by IL-4 in strains. TPM, transcripts per kilobase million. ****q < 0.0001, compared to basal. Numbers indicate fold change by IL-4. (G) Categories of strain-differential IL-4 up-regulated genes based on the differences in basal gene expression. (H) Average log2 gene expression fold change between alleles in hybrid (C57 × SPRET F1) and parental strain under 24-hour IL-4 conditions.
