Figure 5.
Reference tissue and organoid atlases reveal epithelial progenitor and stem cell states during small intestine development
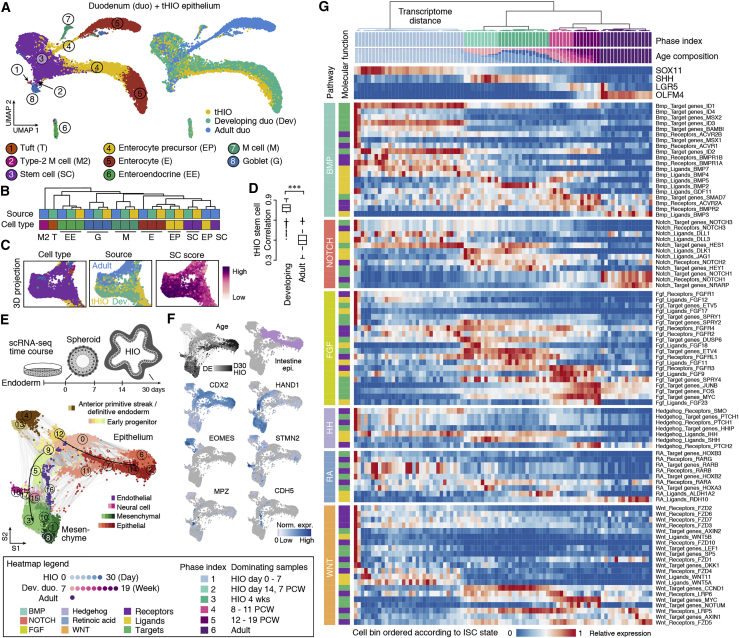
(A) Integrated UMAP of tHIO, developing, and adult duodenum epithelial scRNA-seq datasets, colored by cell type (left) and source (right).
(B) Hierarchical clustering of average transcriptome correlations between cell types of different tissue sources.
(C) Inset of the 3D UMAP, highlighting stem cells colored by cell type (left), source (center), and stem cell score (right).
(D) Boxplots showing the Spearman’s correlation distribution of tHIO stem cells compared with developing or adult stem cells (Wilcoxon rank-sum test, nominal ∗∗∗p < 0.0001).
(E) Schematic of scRNA-seq experiments performed over a time course of in vitro HIO development. SPRING embedding is colored by cell class and shaded by cluster assignment.
(F) HIO time course UMAP colored by time point or marker gene expression. Light purple indicates intestinal epithelial cells.
(G) In vitro HIO, developing, and adult duodenum stem cells ordered by ISC state and colored by phase. Hierarchical clustering of ISC cell bins represents transcriptome distance calculated with ISC development-associated genes. The heatmap shows gene expression changes of signaling pathways during ISC phase transitions.