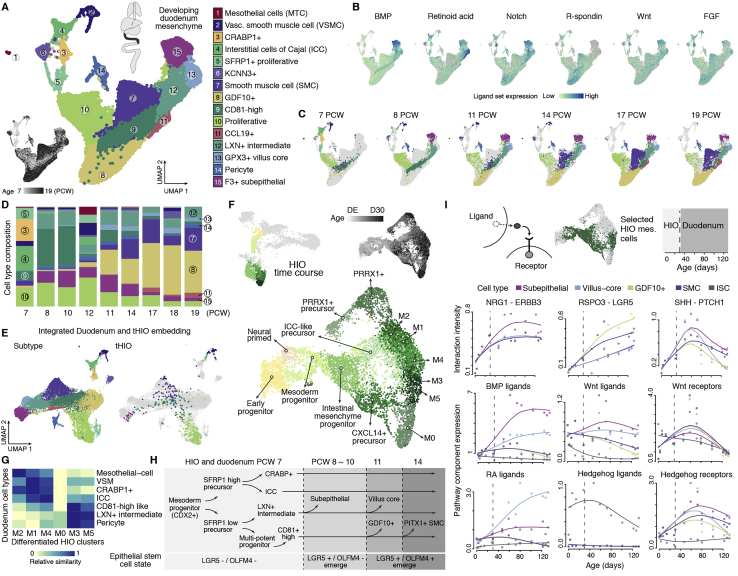
Figure S4.
Integrating HIO and developing duodenum data unveils transient cell states during human intestine mesenchyme development, related to Figures 4 and 5
A) UMAP embedding of mesenchymal cells from developing human early small intestine and duodenum, with cells colored by type (top left) or age (bottom left). Cell type annotations are shown top right. B) Feature plots show the aggregated expression of ligands of each signaling pathway in developing duodenum mesenchyme. C) Time point distribution of mesenchymal cells on the UMAP embedding, with cells colored by type. D) Stacked bar plot shows the proportion of mesenchymal subtypes at different time points in the developing duodenum, with color scheme consistent with panel A. E) Integrated UMAP embedding of developing duodenum and 4-week tHIO intestinal mesenchymal cells. Left, developing duodenum cells in the integrated embedding colored by subtype. Right, tHIO cells colored by the most similar developing duodenum subtype; gray cells represent developing duodenum. F) In vitro HIO time course bi-potent cluster (c9 of HIO time course in Figure 5E) and clusters in the mesenchymal trajectory (c5, c3, c10, c1 and c8) were extracted (top left, clusters from original SPRING embedding are highlighted) and projected to a new UMAP embedding to visualize mesenchymal cell developmental trajectories. Cells were sub-clustered and annotated or colored by age (top right). DE: definitive endoderm. G) Relative transcriptome similarity between differentiated intestinal mesenchymal clusters of HIOs (M1-M5) and mesenchymal subtypes observed in 7 PCW developing duodenum. H) Schematic suggests developmental relationships between duodenum mesenchymal subtypes based on transcriptome similarity and coordinated changes in intestinal epithelial stem cell states. Dashed arrows indicate lower probability. I) Top, schematic shows selected HIO mesenchymal cells as precursors to subepithelial mesenchyme, villus core, GDF10+ mesenchyme and smooth muscle cells. Dashed lines represent separation of in vitro HIO and developing duodenum time points. Bottom, dots represent the interaction intensity between intestinal stem cells and specified mesenchymal subtypes, or signaling pathway component expression of specific cell types at each time point. Lines represent interpolated spline curves. Interaction intensity is quantified as the averaged expression of a ligand-receptor gene pair in intestinal epithelial stem cells and each of the specified mesenchymal subtypes. Pathway signaling component expression is calculated as the sum of normalized expression levels of related genes in each cell type.