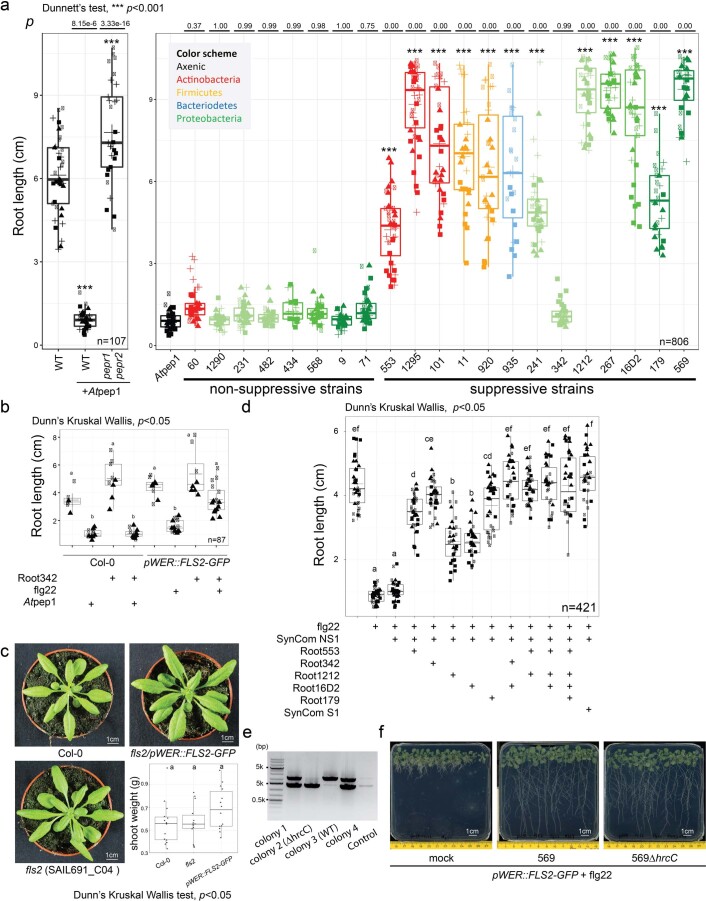
Extended Data Fig. 2. Microbiota members modulate root growth inhibition (RGI) induced by Atpep1 and function quantitatively in community contexts.
a, Root lengths of WT and pepr1pepr2 mutants (left panel) or WT plants (right panel) inoculated with 1 μM Atpep1 alone or with individual strain. b, Caulobacter 342 exhibits differential ability to suppress flg22- or Atpep1-mediated RGI. c, Representative images of WT, fls2 and pWER::FLS2-GFP plants grown in natural Lausanne soil and their corresponding shoot fresh weight without any flg22 treatment. d, Root lengths of pWER::FLS2-GFP plants inoculated with different combination of non-suppressive and suppressive strains in the presence of 1 μM flg22. n=total number of biological samples collected from four (a), two (b), two (c) and three (d) biological replicates. Asterisks indicate statistical significance (two-sided Dunn’s Kruskal Wallis or two-sided Dunnett’s test, p < 0.05). The box plots centre on the median and extend to the 25th and 75th percentiles, and the whiskers extend to the furthest point within the range of 1.5× the interquartile range. e, Gel blot (Source ED Fig. 2e) showing the result of PCR genotyping of 569ΔhrcC generated by homologous recombination. Lane from left to right, GeneRuler 1 kb plus ladder, four individual colonies and control. f, Root lengths of pWER::FLS2-GFP plants coinoculated with flg22 and 569 or 569ΔhrcC.