Fig. 7. Stimulating early PGC-1α activity protects the host.
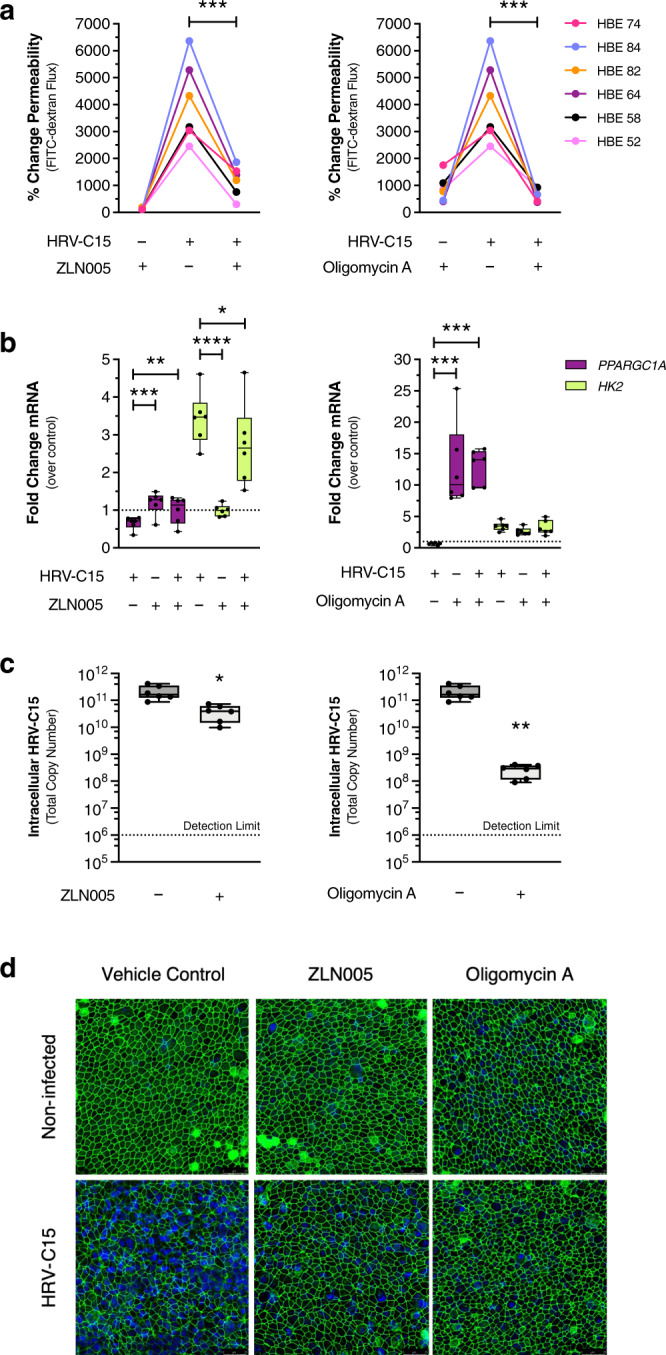
a FITC-dextran (3–5 kDa) permeability assay indicating barrier function recovery at 24 h post infection from early administration of 100 μM ZLN005 (left) or 3 μM oligomycin A (right). Data are represented as individual HBE donor lines and analyzed by one-way ANOVA with Holm–Sidak multiple comparisons: ZLN005 ***p = 0.0002, oligomycin A ***p = 0.0006, n = 6 donors. b Effects of 100 μM ZLN005 (left) or 3 μM oligomycin A (right) on PPARGC1A and HK2 mRNA expression from matched samples derived from (a) at 24 h post infection. Data are represented as boxplots and PPARGC1A and HK2 analyzed separately by one-way ANOVA with Holm–Sidak multiple comparisons. ZLN005: PPARGC1A ***p = 0.0006, **p = 0.009, HK2 ****p < 0.0001, *p = 0.036. Oligomycin A: PPARGC1A ***p = 0.0005, ***p = 0.005 HK2 not significant, n = 6 donors. c Matched intracellular HRV-C15 RNA copy number from ZLN005 or oligomycin A-treated ALI cultures from (a) at 24 h post infection, quantified by RT-PCR. Dashed line represents RT-PCR detection limit. There was no detectable HRV-C15 in noninfected ALI cultures. Data are represented as boxplots and analyzed by two-tailed paired t test: ZLN005 *p = 0.011, oligomycin A **p = 0.009, n = 6 donors. d Representative immunofluorescence images of ZO-1 (green) and nuclei (blue) from a matched experiment in (a) at 24 h post infection. Representative confocal images were obtained from three independent experiments performed using ALI cultures derived from three different lung donors and selected from ten fields per condition per donor, n = 3 donors. Scale bar, 25 μm. Data represented as boxplots indicate the median (center line), upper and lower box bounds (IQR = first and third quartiles), and whiskers (min and max values), with individual donor data points superimposed onto the boxplot.
