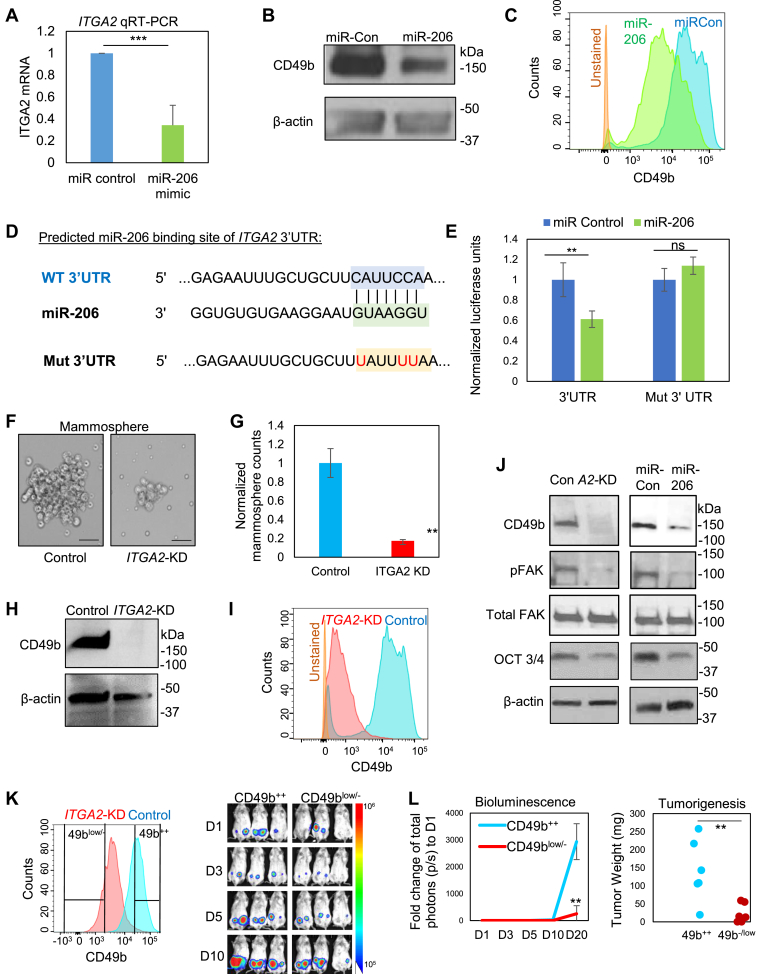
Figure 2.
ITGA2 is a target of miR-206 and promotes mammosphere formation and stemness factor expression. (A–B) ITGA2 mRNA (top panel) and protein (bottom panel) levels are reduced by transfected miR-206 in MDA-MB-231 cells, measured by qRT-PCR and immunoblotting, respectively. ***t-test P < 0.001. Error bars represent S.D. values. (C) CD49b surface protein expression inhibited by miR-206 in MDA-MB-231 cells, evaluated by flow cytometry. (D) Predicted binding sites between the complementary sequences of ITGA2 3’UTR (WT) and miR-206. The bottom line shows the mutated C to T (U, in red) within the interaction site of 3’UTR. (E) 3’UTR luciferase assay shows direct regulation of ITGA2 by miR-206, n = 3, **t-test P = 0.005. (F-G) Images (F) and quantified counts (G) of breast cancer cell-derived mammospheres upon siITGA2 knockdown. **t-test P < 0.01. Error bars represent S.D. values. Scale bars = 25 μm (H-I) siITGA2 transfection depletes CD49b protein expression, measured by immunoblotting (H) and flow cytometry analyses (I). (J) Immunoblots to detect reduced expression of CD49b, phosphorylated FAK (pFAK), and OCT3/4 levels without affecting total FAK levels upon ITGA2 KD or miR-206 upregulation. (K) Left panel: Flow histogram of CD49b expression in control and siITGA2-transfected cells and gated sorting of CD49b++ and CD49blow/- populations, respectively. Right panel: Bioluminescent images of CD49b++ and CD49blow/- implants of 100–1000 cells from day 1–10 (D1-D10). (L) Left panel: Fold change of tumor burden (total photons) to Day 1 (D1) from D1-D20. ** t-test P = 0.007 on D20. Error bars represent S.D. values. Right panel: Weight of tumors derived from CD49b++ and CD49b−/low cells, orthotopically implanted at 100–1000 cells (t-test **P = 0.008).