Abstract
Background
Pseudomonas aeruginosa (P. aeruginosa) is an opportunistic pathogen responsible for burn-wound infection. High incidence, infection severity and increasing resistance characterize P. aeruginosa -induced burn infection.
Purpose
To estimate quorum-sensing (QS)-dependent virulence factors of P. aeruginosa isolates from burn wounds and correlate it to the presence of QS genes.
Methods
A cross-sectional descriptive study included 50 P . aeruginosa isolates from burn patients in Mansoura University Plastic and Burn Hospital, Egypt. Antibiotic sensitivity tests were done. All isolates were tested for their ability to produce biofilm using a micro-titration assay method. Protease, pyocyanin and rhamnolipid virulence factors were determined using skimmed milk agar, King’s A medium and CTAB agar test, respectively. The identity of QS lasR and rhlR genes was confirmed using PCR.
Results
In total, 86 % of isolates had proteolytic activity. Production of pyocyanin pigment was manifested in 66 % of isolates. Altogether, 76 % of isolates were rhamnolipid producers. Biofilm formation was detected in 96 % of isolates. QS lasR and rhlR genes were harboured by nearly all isolates except three isolates were negative for both lasR and rhlR genes and two isolates were positive for lasR gene and negative for rhlR gene. Forty-nine isolates were considered as extremely QS-proficient strains as they produced QS-dependent virulence factors. In contrast, one isolate was a QS deficient strain.
Conclusions
QS affects P. aeruginosa virulence-factor production and biofilm in burn wounds. Isolates containing lasR and rhlR seem to be a crucial regulator of virulence factors and biofilm formation in P. aeruginosa whereas the lasR gene positively regulates biofilm formation, proteolytic activity, pyocyanin production and rhamnolipid biosurfactant synthesis. The QS regulatory RhlR gene affects protease and rhamnolipid production positively.
Keywords: Pseudomonas aeruginosa, virulence, QS, biofilm
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a Gram-negative bacillus, which is considered one of the most important opportunistic pathogens, and establishes itself in vulnerable immunocompromised patients causing urinary tract infections, respiratory system infections, bacteremia and burn-wound infections. In patients with severe burn wounds, it is difficult to treat and can cause high mortality [1]. However, this bacillus can be found on the skin, throat and stool of non-hospitalized patients [2].
Multiply resistant organisms, including Pseudomonas and Acinetobacter , are the leading causes of death from infection after burns [3]. Numerous P. aeruginosa virulence factors contribute to the pathogenesis of burn-wound infection such as pili, flagella, biofilm formation besides the protease, pyocyanin and rhamnolipid extracellular virulence factors. Due to a range of mechanisms for virulence and antibiotic resistance, understanding the regulatory mechanisms regulating the expression of virulence genes is critical for the development of alternative therapeutic approaches in order to monitor and prevent these pseudomonal infections [4]. The creation of biofilms and the bacterial activity have been regulated by quorum sensing (QS) and often render Pseudomonas infections difficult to eradicate [5].
QS refers to the ability of a bacterium to sense information from other cells in the population when they reach a critical concentration (i.e. a quorum) and communicates with them to regulate the production of variable virulence factors using small, signalling molecules called autoinducers [6]. P. aeruginosa contains two main QS systems, las and rhl. Each system consists of two components, the autoinducer synthases (lasI and rhlI, respectively) and their cognate transcriptional regulators (lasR and rhlR, respectively). P. aeruginosa also possesses a third QS system called Pseudomonas quinolone signal (PQS) and a recently identified fourth QS system termed as the integrated QS (IQS) system [7].
These systems control expression of many extracellular virulence factors that play crucial roles in colonizing host tissues and are regulated by the las system and the rhl system [8]
Our aim was to estimate QS-dependent virulence factors as protease, pyocyanin, rhamnolipid and biofilm-producing ability of P. aeruginosa isolates from burn wounds and correlate it to the presence of QS genes.
Methods
This is a cross-sectional descriptive study of 100 patients with burns who were admitted to the plastic surgery and burn centre at Mansoura University Hospital (MUH), from September 2017 to August 2018, included 65 males and 35 females with ages ranging from 2 to 54 years (mean±sd 27.74±12.93).
Swabs were collected using Levine’s technique by rotating manoeuvre over 1 cm2 area of the wound with sufficient pressure to extract fluid from within the wound tissue [9] and were transported aseptically in Stuart’s transport media. Primary plating of all samples was performed quantitatively on nutrient, blood and MacConkey’s agar, incubated aerobically at 37 °C. The infected wound was defined as those containing 1×105 organisms or more per swab [9]. The colonial forms were observed, and the characteristic odour of the organism was detected. Colonies were examined for pigment production on cetrimide agar at 42 °C. However, some strains, proved to be P. aeruginosa but were not fluorescent. Gram-stained smears from suspected colonies showed non-sporeforming Gram-negative bacilli.
Colonies were reexamined using different biochemical tests; oxidase-positive colonies became dark blue within a few seconds, catalase-positive isolates detected by the presence of immediate effervescence, Fig. 1.
Fig. 1.

Different biochemical reactions of P. aeruginosa isolates (KIA=Kligler iron agar medium, MIO=motility indole ornithine reaction).
The tested organism was subcultured and incubated on Kligler iron agar (KIA) medium; it was examined for the colour change caused by sugar fermentation, gas and H₂S production, P. aeruginosa isolates are neither glucose nor lactose fermenter, detected by pink red slant and butt with no gas or H₂S production. Motility indole ornithine (MIO) reaction allows detection of the organism’s motility along the stab line of inoculation, Fig. 1.
n’s citrate medium was inoculated by the tested organism; P. aeruginosa isolates produce positive citrate utilization test, the presence of alkaline product is proved by the changing colour into blue. Speaking of which, the test strain was streaked on urea agar slant where P. aeruginosa isolates are either urease positive or urease negative Fig. 1.
Furthermore, the automatic VITEK 2 (bioMérieux) was used to identify the bacterial species, antibiogram profile of the isolates were deduced by disc diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) [10].
Biofilm production of P. aeruginosa isolates was assessed by using a micro-titration assay method previously described by Stepanović et al. [11]. Biofilm-forming capacity was calculated at OD 492 nm using a microtitre plate reader. The taken value was the mean of three readings for each clinical isolate. The tested isolates were classified into three categories according to the value of OD 492 nm as strong, moderate and weak biofilm adherence.
The assessment of P. aeruginosa proteolytic activity was performed using skimmed milk agar. The previously identified P. aeruginosa strains were streaked on this media, the clearance of skimmed milk occurred by hydrolysis of casein (a primary milk protein) [12]. Pyocyanin quantification was executed using King’s A broth media that enhances pyocyanin pigment production. Pyocyanin was extracted using chloroform and its absorbance was measured in the acidic form. The pyocyanin amount throughout the supernatant fractions of P. aeruginosa isolates is evaluated by the absorbance value at 520 nm. A higher absorption value implies higher production of pigments [13].
Rhamnolipid was assayed using cetyl trimethyl ammonium bromide (CTAB) assay method using M9 minimal media. Overall, 50 µl of supernatant from 24 h incubated bacterial broth at 37 °C were inoculated in performed agar wells, then the plates were incubated for 24–48 h at 37 °C [14].
PCR analysis was carried out using internal primers of QS genes (lasR and rhlR) as described previously by Cotar et al. [15], Table 1. Bacterial genomic DNA was extracted using aGeneJET Genomic DNA purification kits (Thermo Fisher Scientific, EU) from all phenotypically and biochemically tested strains, as well as from the reference positive control strain P. aeruginosa (PAO1). By the use of DNA thermal cycler, PCR reaction was carried out in a 25 µl total volume. The cycling conditions incorporated initial denaturation followed by 35 cycles at 94 °C for 30 s for DNA denaturation, followed by annealing temperature at 50 °C for 30 s, extension at 72 °C for 2 min and finally the process was ended by final extension at 72 °C for 5 min [15].
Table 1.
Primers that were used for detection of lasR and rhlR genes by PCR [15]
|
Gene type |
Primer direction |
Primer sequence (5′ to 3′) |
Amplicon size (bp) |
|---|---|---|---|
|
LasR |
Fw |
5′-ATGGCCTTGGTTGACGGTT-3′ |
725 |
|
Rev |
5′-GCAAGATCAGAGAGTAATAAGACCCA-3′ |
||
|
RhlR |
Fw |
5′-CAATGAGGAATGACGGAGGC-3′ |
730 |
|
|
Rev |
5′-GCTTCAGATGAGGCCCAGC-3′ |
Following amplification, aliquots (10 µl) were removed from each reaction mixture and examined by means of gel electrophoresis composed of 1.5 % agarose in TBE buffer (40 mM Tris, 20 mM boric acid, 1 mM EDTA, pH 8.3). Then, the gels were Image graphed under UV lights and the DNA bands were visualized by ethidium bromide staining [15]. The PCR products were estimated by comparison with the 100 bp DNA ladder molecular size markers.
Statistical analysis
Data were analysed using the Statistical Package of Social Science (SPSS) programme for Windows (Standard version 21). The normality of data was first tested with one-sample Kolmogorov–Smirnov test. Qualitative data were described using number and percent. Association between categorical variables was tested using Fischer's exact test when expected cell count was less than 5. Continuous variables were presented as mean±sd for parametric data and median (min-max) for non-parametric data. The two groups were compared with Student's t-test for parametric data and Mann–Whitney test for non-parametric data. Probability levels≤0.05 were considered statistically significant.
Results
In the present study, 110 isolates from 100 patients with burns were collected. P. aeruginosa was the most common pathogen that was isolated from 50 swabs (45 %) followed by Klebsiella pneumoniae isolated from 26 swabs (24 %), Staphylococcus aureus isolated from 12 swabs (11 %), Escherichia coli isolated from nine swabs (8 %) and Proteus mirabilis isolated from seven swabs (6 %), Table 2.
Table 2.
Bacterial isolates from burn wounds
|
Organism |
No. [Total no.=110] |
Percentage |
|---|---|---|
|
50 |
45 % |
|
|
26 |
24 % |
|
|
12 [6 MRSA and 6 MSSA] |
11 % |
|
|
9 |
8 % |
|
|
7 |
6 % |
|
|
Enterococci |
3 |
3 % |
|
Citrobacter species |
2 |
2 % |
|
Streptococcal pyogenes |
1 |
1 % |
Identification of all the collected Pseudomonas specimens was confirmed. The physiological properties of all isolates proved their ability to produce pigment, grow at 42 °C, being oxidase positive and catalase positive. Different biochemical tests were used to identify P. aeruginosa isolates as KIA where they were sugar non-fermenters. MIO test showed that all isolates are motile. Isolates of P. aeruginosa showed their ability to utilize citrate and variable urease enzyme production.
The isolates were reported to be sensitive, intermediate and resistant from the respective interpretation charts recommended by CLSI [10] where the highest sensitivity of P. aeruginosa isolates was to colistin (100 %) followed by polymyxin B (96 %). Meanwhile, the least sensitivity was recorded against ceftazidime (6%). Forty-six (92 %) of isolates were MDR as displayed in Table 3.
Table 3.
The antibiotic susceptibility pattern of P. aeruginosa isolates
|
Sensitive |
Resistant |
|||
|---|---|---|---|---|
|
Number |
Percentage % |
Number |
Percentage % |
|
|
Piperacillin (PRL) |
4 |
8 % |
46 |
92 % |
|
Piperacillin-tazobactam (TPZ) |
23 |
46 % |
27 |
54 % |
|
Ceftazidime (CAZ) |
3 |
6 % |
47 |
94 % |
|
Cefepime (FEP) |
4 |
8 % |
46 |
92 % |
|
Aztreonam (ATM) |
4 |
8 % |
46 |
92 % |
|
Imipenem (IMP) |
21 |
42 % |
29 |
58 % |
|
Colistin (CT) |
50 |
100 % |
0 |
0 % |
|
Polymyxin B (POB) |
48 |
96 % |
2 |
4 % |
|
Gentamycin (CN) |
8 |
16 % |
42 |
84 % |
|
Tobramycin (TOB) |
13 |
26 % |
37 |
74 % |
|
Amikacin (AK) |
8 |
16 % |
42 |
84 % |
|
Ciprofloxacin (CIP) |
16 |
32 % |
34 |
68 % |
|
Levofloxacin (LEV) |
19 |
8 % |
46 |
92 % |
|
MDR Yes No |
46(92 %) 4(8 %) |
|||
Biofilm-producing ability was detected by way of strain that was twice that of the negative control OD, considered positive. Seven (14 %) P. aeruginosa isolates exhibited strong adherence as they produced biofilm with OD 492 nm ≥0.272 compared with PAO1 that was used as a positive control and had an OD 0.374. Nineteen (38 %) isolates were moderately adherent as they produced biofilm ranged between 0.136<ODT≤0.272, the remaining isolates (44 %) with biofilm ranged between 0.068<ODT≤0.136 reflected weak producers. However, two (4 %) isolates were non-biofilm producers, Figs. 2 and 3.
Fig. 2.
The percentage of degrees of biofilm producers among P. aeruginosa isolates.
Fig. 3.
Micro-titration plate showing different grades of P. aeruginosa biofilm.
The majority of P. aeruginosa isolates 43 (86 %) had proteolytic activity whereas 7 (14 %) showed no proteolytic activity, Figs 4 and 5. Quantification of pyocyanin production by the isolates occurred as described before by Essar et al. [13]. Thirty three (66 %) P. aeruginosa isolates produced pyocyanin pigment and 17 (34 %) lack the ability for pyocyanin production, five isolates produced a high amount of pyocyanin ≥8.365 µg ml−1 compared with the positive control PAO1 and 28 isolates produced a moderate amount of pyocyanin ranged between the negative control (double mutant PAO-JP2) (1.536 µg ml−1) and positive control PAO1 (8.365 µg ml−1), Fig. 6. An evidence of rhamnolipid production was detected in 38 (76 %) isolates that were rhamnolipid producers, on the other hand, the remaining 12 (24 %) isolates were categorized as rhamnolipid non-producers, Fig. 7.
Fig. 4.
Positive protease on skimmed milk agar.
Fig. 5.
Negative protease on skimmed milk agar.
Fig. 6.
Pyocyanin pigment detection.
Fig. 7.
Qualitative detection of rhamnolipid produced by P. aeruginosa isolates using cetyl trimethyl ammonium bromide (CTAB) assay method.
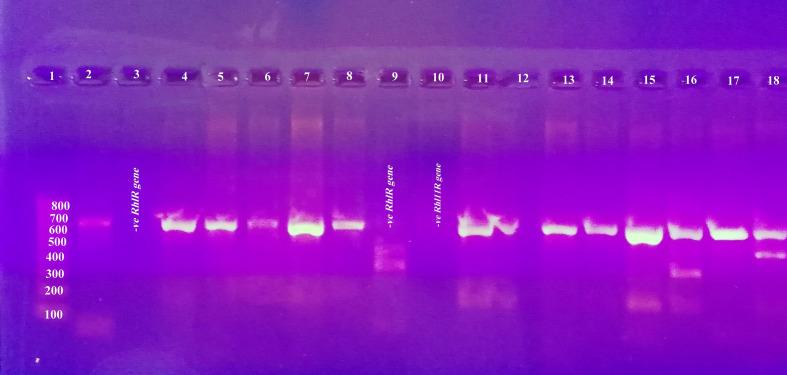
PCR detection of lasR and rhlR genes showed that they were harboured by nearly all isolates except three isolates with amplicon sizes of 725 and 730 bp, respectively. Regarding lasR, it was detected in 47 (94 %) out of 50 P . aeruginosa isolates collected as only three isolates did not harbour the gene, Fig. 8. There was a statistically significant difference between lasR-positive isolates and lasR-negative isolates regarding biofilm production (weak, moderate and strong) (P value=0.031), Table 4. Also, there was a highly statistically significant difference between lasR-positive isolates and lasR-negative isolates regarding protease production (P value=<0.001). Once more, there was a statistically significant difference between lasR-positive isolates and lasR-negative isolates regarding pyocyanin production (P value=0.035). Equally there was a statistically significant difference between lasR-positive isolates and lasR-negative isolates regarding rhamnolipid production (P value=0.011).
Fig. 8.
Agarose gel electrophoresis for lasR amplicons in P. aeruginosa isolates.
Table 4.
LasR gene and biofilm formation in P. aeruginosa isolates
|
Positive lasR gene (n=47) |
Negative lasR gene (n=3) |
P value |
|||
|---|---|---|---|---|---|
|
No |
% |
No |
% |
||
|
Biofilm formation |
0.031* |
||||
|
Non-biofilm former |
1 |
2.1 |
1 |
33.3 |
|
|
Weak-biofilm former |
20 |
42.6 |
2 |
66.7 |
|
|
Moderate-biofilm former |
19 |
40.4 |
0 |
0.0 |
|
|
Strong-biofilm former |
7 |
14.9 |
0 |
0.0 |
|
The rhlR gene was detected in 45 (90 %) out of 50 P. aeruginosa isolates collected, while only five (10 %) isolates did not harbour the gene, Fig. 9 . There was no statistically significant difference between rhlR-positive isolates and rhlR-negative isolates regarding biofilm production (weak, moderate and strong) (P value=0.163), Table 5. Identically, there was statistically significant difference between rhlR-positive isolates and rhlR-negative isolates regarding protease production (P value=0.016). Dissimilarly, there was no statistically significant difference between rhlR-positive isolates and rhlR-negative isolates regarding pyocyanin production (P value=0.321). There was a highly statistically significant difference between rhlR-positive isolates and rhlR-negative isolates regarding rhamnolipid production (P value=<0.001).
Fig. 9.
Agarose gel electrophoresis for rhlR amplicons in P. aeruginosa isolates.
Table 5.
RhlR gene and biofilm formation in P. aeruginosa isolates
|
Resistant |
Positive rhlR gene (n=45) |
Negative rhlR gene (n=5) |
P value |
||
|---|---|---|---|---|---|
|
No |
% |
No |
% |
||
|
Biofilm formation |
0.163 |
||||
|
Non-biofilm former |
1 |
2.2 |
1 |
20.0 |
|
|
Weak-biofilm former |
19 |
42.2 |
3 |
60.0 |
|
|
Moderate-biofilm former |
18 |
40.0 |
1 |
20.0 |
|
|
Strong-biofilm former |
7 |
15.6 |
0 |
0.0 |
|
Forty-nine (98 %) isolates displayed the QS-proficient phenotype and one (2 %) isolate was found to be lacking all tested virulence factors. Variable production of virulence factors is shown in Table 6.
Table 6.
Virulence factors, QS lasR and rhlR genes in P. aeruginosa isolates
|
Virulence factors |
QS gene |
|||||
|---|---|---|---|---|---|---|
|
No. of isolates |
Biofilm |
Total protease |
Pyocyanin |
Rhamnolipid |
LasR gene |
RhlR gene |
|
8 |
Weak producer |
+ |
+ |
+ |
+ |
+ |
|
12 |
Moderate producer |
+ |
+ |
+ |
+ |
+ |
|
5 |
Strong producer |
+ |
+ |
+ |
+ |
+ |
|
2 |
Weak producer |
− |
+ |
+ |
+ |
+ |
|
1 |
Weak producer |
+ |
+ |
− |
+ |
+ |
|
1 |
Weak producer |
+ |
+ |
− |
+ |
− |
|
1 |
Moderate producer |
+ |
+ |
− |
+ |
+ |
|
1 |
Moderate producer |
+ |
+ |
− |
+ |
− |
|
1 |
Strong producer |
+ |
+ |
− |
+ |
+ |
|
1 |
Non-biofilm producer |
+ |
+ |
− |
+ |
+ |
|
2 |
Weak producer |
+ |
− |
− |
+ |
+ |
|
1 |
Moderate producer |
+ |
− |
− |
+ |
+ |
|
5 |
Weak producer |
+ |
− |
+ |
+ |
+ |
|
4 |
Moderate producer |
+ |
− |
+ |
+ |
+ |
|
1 |
Weak producer |
− |
− |
+ |
+ |
+ |
|
1 |
Strong producer |
− |
− |
+ |
+ |
+ |
|
2 |
Weak producer |
− |
− |
− |
− |
− |
|
1 |
Non-biofilm producer |
− |
− |
− |
− |
− |
|
Total=50 | ||||||
Discussion
P. aeruginosa is an opportunistic human pathogen capable of causing a wide array of extracellular virulence factors besides antibiotic resistance that continues to evolve [16]. These virulence factors regulated by QS help bacteria evade the host immune system. Strategies for QS disruption have become an attractive target for the development of new therapeutic measures.
According to our antibiogram results, the antibiotic susceptibility pattern of isolated P. aeruginosa showed the highest sensitivity of P. aeruginosa isolates was to colistin (100 %) followed by polymyxin B (96 %) and the least sensitivity was recorded against ceftazidime (6 %). These results showed that P. aeruginosa has become more resistant to drugs that were thought to be efficient previously. This was in accordance with another study carried out by Choudhary et al. [17] that revealed the resistance of P. aeruginosa was alarmingly high, where the sensitivity for piperacillin-tazobactam was 38.9 %, amikacin 25 %, imipenem 23.4 %, meropenem 19 %, aztreonam 19.4 %, ceftazidime 8.8 % and for cefepime 8.3 %. In disagreement to this study, another study by Revathi et al. [18] reported P. aeruginosa isolates were susceptible to commonly used antibiotics. This may be due to increased emergency of resistant Pseudomonas strains due to increased use of antibiotics to treat infections.
In this study, P. aeruginosa isolates showed biofilm-forming capacity. Out of 50 P. aeruginosa isolates, 48 (96 %) isolates were biofilm producers [7 (14 %) isolates were strong biofilm producers, 19 (38 %) were moderate biofilm producers and 22 (44 %) were weak biofilm producers], whereas two (4 %) isolates were non-biofilm producers. Our results were consistent with Jabalameli et al. [19] who reported a high percentage of P. aeruginosa biofilm formers, in which positive P. aeruginosa isolates for biofilm were about 96 % [47, 26 and 22.9 % were strong, moderate and weak biofilm producers, respectively]. In disagreement with our results, Heydari and Eftekhar reported biofilm formation in only 43.5 % of P. aeruginosa isolates (66.7 % strong and 33.3 % weak biofilm formers) [20].
Proteases enzymes participate significantly in the P. aeruginosa pathogenesis; degrade host tissues and enhance the bacterial growth and invasiveness in burnt patients [21]. In the current study, 50 P. aeruginosa isolates were screened for total protease production; 43 (86 %) isolates were positive protease while seven (14 %) isolates were negative protease. Our results were in agreement with Khalil et al. who reported a high percentage (95 %) of protease production in P. aeruginosa isolates from burn wounds [22]. This promotes the opinion that says in acute P. aeruginosa infections, protease activity is seen more and reduces when infection become chronic [23].
Pyocyanin has the ability to arrest different micro-organism’s electron transport chain and to exhibit a wide range of cell damage and antimicrobial activity [24]. We assessed the ability of P. aeruginosa isolates to produce pyocyanin and we detected a high rate of pyocyanin production, where 33 (66 %) isolates were positive pyocyanin producers and 17 (34 %) isolates were negative pyocyanin producers. This result corresponds to the study conducted by Khadim and Marjani [25] who proved high pyocyanin production in 35 % of P. aeruginosa isolates of burn patients.
Rhamnolipids influence the biofilm architecture [26]. Rhamnolipid was found positive in 38 (76 %) of the P. aeruginosa isolates in our study. This is similar to a study done by Khalil et al. [22] who documented 80 % positive rhamnolipid in P. aeruginosa isolates collected from burn patients.
Las system is involved in early stages of biofilm formation where O’Toole and Kolter [27] found that mutation in the las synthase gene resulted in defective, uniform, flat and undifferentiated biofilms. The rhl system shares biofilm maturation through deposition of rhamnolipid [28]. Protease and elastase are regulated by the las system, however pyocyanin and rhamnolipid are regulated by the rhl system [29].
Deficient lasR strains were found to be less virulent [30]. In our study of QS lasR and rhlR genes, the lasR gene was detected in 94 % of P. aeruginosa isolates. This result is consistent with El-Khashaab et al. [31] who reported 94.3 % of P. aeruginosa isolates that were positive for the lasR gene.
In the current study, all biofilm-producer isolates were found to harbour the lasR gene with statistically significant difference between lasR-positive isolates and lasR-negative isolates regarding biofilm production (P value=0.031). This is similar to data published by Li et al. [32].
C12-HSL/lasR regulates the production of different virulence factors including elastase (lasB), toxA and lasA protease [33]. Our results revealed a highly statistically significant difference between lasR-positive isolates and lasR-negative isolates regarding protease production (P value=<0.001).
Certainly, pyocyanin secretion is under the control of the rhl systems. As well, PQS controls rhl-dependent virulence-factor production such as pyocyanin [34], as regards lasR, the study done by Abou shleib and his colleagues found that there was no statistical significant value between pyocyanin production and the studied lasR gene [35]. In contrast, in our study we found a statistically significant difference between lasR-positive isolates and lasR-negative isolates regarding pyocyanin production (P value=0.035).
Rhamnolipid is under rhl system control as well as PQS [ 34 ]. We found a statistically significant difference between lasR-positive isolates and lasR-negative isolates regarding rhamnolipid production (P value=0.011). This is in agreement with a study done by Henkel et al. [36] who reported a role of AHL in regulating rhamnolipid production. In addition, las system controls the rhl system.
In this study, there was no statistically significant difference between rhlR-positive isolates and rhlR-negative isolates regarding biofilm production (P value=0.163), which is dissimilar to the study conducted by Li et al. [32] who reported positive statistically significant value between rhlR expression and biofilm formation only on day 14 (P<0.05).
In this study, there was statistically significant difference between rhlR-positive isolates and rhlR-negative isolates regarding protease production (P value=0.016). In agreement to our results, Van Delden and Iglewski [37] reported that an intact rhl system is required for the restoration of elastase in the absence of lasR. But, we were unable to use this approach to understand what happened with the rhlR, lasI or rhlI mutant, since such mutants still produce enough elastase.
Pyocyanin is a rhl-related virulence factor. In many studies conducted by Lee and Zhang [34] and Reuter et al. [38], they used the production of pyocyanin by P. aeruginosa as an indicator for rhl and PQS system activity. In the present study there was no statistically significant difference between rhlR-positive isolates and rhlR-negative isolates regarding pyocyanin production (P value=0.321).
Another exoproduct controlled primarily by rhlR is rhamnolipid where its production relies on several environmental and nutritional factors, including nitrogen and iron diminution, pH and temperature [39]. In our results there was a highly statistically significant difference between rhlR-positive isolates and rhlR-negative isolates regarding rhamnolipid production (P value=<0.001).
Schaber et al. [40] reported one QS-deficient clinical isolate that caused a wound infection even in the absence of all tested virulence factors. QS-dependent phenotypes may be positive for all four QS genes as observed by Karatuna and Yagci [41]. However, Schaber and colleagues isolated P. aeruginosa strain with no elastase or pyocyanin activity, although having all QS genes [40].
Our study has some limitations, such as small sample size for survey, which can be validated by further large-scale studies. Methodology-related limitations included the swab sampling, which could be replaced by wound biopsy to provide definite evidence of burn-wound infection, and studying of some phenotypic virulence factors and only 2 QS genes ignoring the other genes.
Conclusions
QS affects P. aeruginosa virulence-factor production and biofilm in burn wounds. QS-deficient isolates failed to amplify lasR and rhlR genes however it is still capable of causing infections by other mechanisms. Isolates containing lasR and rhlR seem to be a crucial regulator of virulence factors and biofilm formation in P. aeruginosa whereas the lasR gene positively regulates biofilm formation, proteolytic activity, pyocyanin production and rhamnolipid biosurfactant synthesis. The QS regulatory RhlR gene affects protease and rhamnolipid production positively.
Funding information
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The protocol of this study was accepted by Institutional Review Board (IRB), Faculty of Medicine, Mansoura University; code number: R/16.12.81.
Footnotes
Abbreviations: CTAB, cetyl trimethyl ammonium bromide; KIA, kligler iron agar medium; MIO, motility indole ornithine reaction; OD, optic density; QS, quorum sensing.
References
- 1.Streeter K, Katouli M, Genecology Research Centre, Faculty of Science, Health, Education and Engineering, University of the Sunshine Coast, Maroochydore DC, Queensland, Australia Pseudomonas aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. Infect Epidemiol Microbiol. 2016;2:25–32. doi: 10.18869/modares.iem.2.1.25. [DOI] [Google Scholar]
- 2.Rossolini GM, Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa . Clinical Microbiology and Infection. 2005;11:17–32. doi: 10.1111/j.1469-0691.2005.01161.x. [DOI] [PubMed] [Google Scholar]
- 3.Norbury W, Herndon DN, Tanksley J, Jeschke MG, Finnerty CC. Infection in burns. Surg Infect. 2016;17:250–255. doi: 10.1089/sur.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Japoni A, Farshad S. Alborzi A. Pseudomonas aeruginosa: burn infection, treatment and antibacterial resistance. Iran Red Crescent Med J. 2009;11:244–253. [Google Scholar]
- 5.Taylor PK, Yeung ATY, Hancock REW. Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol. 2014;191:121–130. doi: 10.1016/j.jbiotec.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarabhai S, Kaur A, Capalash N, Sharma P. Quorum sensing in Pseudomonas aeruginosa. In: Kahlon RS, editor. Mechanism and Regulation of Virulence. Pseudomonas Molecular and Applied Biology. 1st ed., chapter 6. Springer International Publishing; 2016. pp. 231–256. editor. [Google Scholar]
- 8.Luo J, Kong J-L, Dong B-Y, Huang H, Wang K, et al. Baicalein attenuates the quorum sensing-controlled virulence factors of Pseudomonas aeruginosa and relieves the inflammatory response in P. aeruginosa-infected macrophages by downregulating the MAPK and NFκB signal-transduction pathways. Drug Des Devel Ther. 2016;10:183–203. doi: 10.2147/DDDT.S97221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner SE, Frantz RA, Saltzman CL, Hillis SL, Park H, et al. Diagnostic validity of three swab techniques for identifying chronic wound infection. Wound Repair Regen. 2006;14:548–557. doi: 10.1111/j.1743-6109.2006.00162.x. [DOI] [PubMed] [Google Scholar]
- 10.CLSI Performance standards for antimicrobial Disk Susceptibility tests. 26th ed. CLSI standards M100S. Wayne, PA: Clinical and laboratory standards institute, Pennsylvania; 2016. [Google Scholar]
- 11.Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 12.Brown MR, Foster JH. A simple diagnostic milk medium for Pseudomonas aeruginosa . J Clin Pathol. 1970;23:172–177. doi: 10.1136/jcp.23.2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/JB.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinzon NM, Ju L-K. Improved detection of rhamnolipid production using agar plates containing methylene blue and cetyl trimethylammonium bromide. Biotechnol Lett. 2009;31:1583–1588. doi: 10.1007/s10529-009-0049-7. [DOI] [PubMed] [Google Scholar]
- 15.Cotar A, I, Dinu S O RIN, Chifiriuc MC, Banu O T I LIA, Iordache C, et al. Screening of molecular markers of quorum sensing in Pseudomonas aeruginosa strains isolated from clinical infections. Rom Biotechnol Lett. 2008;13:3765–3771. [Google Scholar]
- 16.Maurice NM, Bedi B, Sadikot RT. Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am J Respir Cell Mol Biol. 2018;58:428–439. doi: 10.1165/rcmb.2017-0321TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhary V, Pal N, Hooja S. Prevalence and antibiotic resistance pattern of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from clinical specimens in a tertiary care hospital. J Mahatma Gandhi Inst Med Sci. 2019;24:19–22. [Google Scholar]
- 18.Revathi G, Puri J, Jain BK. Bacteriology of burns. Burns. 1998;24:347–349. doi: 10.1016/S0305-4179(98)00009-6. [DOI] [PubMed] [Google Scholar]
- 19.Jabalameli F, Mirsalehian A, Khoramian B, Aligholi M, Khoramrooz SS, et al. Evaluation of biofilm production and characterization of genes encoding type III secretion system among Pseudomonas aeruginosa isolated from burn patients. Burns. 2012;38:1192–1197. doi: 10.1016/j.burns.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 20.Heydari S, Eftekhar F. Biofilm formation and β-lactamase production in burn isolates of Pseudomonas aeruginosa . Jundishapur J Microbiol. 2015;8:e15514. doi: 10.5812/jjm.15514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed AA, Salih FA. Low concentrations of local honey modulate Exotoxin A expression, and quorum sensing related virulence in drug-resistant Pseudomonas aeruginosa recovered from infected burn wounds. Iran J Basic Med Sci. 2019;22:568–575. doi: 10.22038/ijbms.2019.33077.7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khalil MAEF, Ibrahim Sonbol F, Mohamed AFB, Ali SS, Sonbol F., I Comparative study of virulence factors among ESβL-producing and nonproducing Pseudomonas aeruginosa clinical isolates. Turk J Med Sci. 2015;45:60–69. doi: 10.3906/sag-1311-102. [DOI] [PubMed] [Google Scholar]
- 23.Macin S, Akarca M, Sener B, Akyon Y. Comparison of virulence factors and antibiotic resistance of Pseudomonas aeruginosa strains isolated from patients with and without cystic fibrosis. Revista Romana de Medicina de Laborator. 2017;25:327–334. doi: 10.1515/rrlm-2017-0027. [DOI] [Google Scholar]
- 24.Laxmi M, Bhat SG. Characterization of pyocyanin with radical scavenging and antibiofilm properties isolated from Pseudomonas aeruginosa strain BTRY1. 3 Biotech. 2016;6:27–32. doi: 10.1007/s13205-015-0350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khadim MM. Marjani M F A. pyocyanin and biofilm formation in Pseudomonas aeruginosa isolated from burn infections in Baghdad, Iraq. Biological. 2019;12:131–145. [Google Scholar]
- 26.Nickzad A, Déziel E. The involvement of rhamnolipids in microbial cell adhesion and biofilm development - an approach for control? Lett Appl Microbiol. 2014;58:447–453. doi: 10.1111/lam.12211. [DOI] [PubMed] [Google Scholar]
- 27.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 28.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H, Bandara R, Conibear TCR, Thuruthyil SJ, Rice SA, et al. Pseudomonas aeruginosa with lasI quorum-sensing deficiency during corneal infection. Invest Ophthalmol Vis Sci. 2004;45:1897–1903. doi: 10.1167/iovs.03-0980. [DOI] [PubMed] [Google Scholar]
- 31.El-Khashaab TH, Erfan DM, Kamal A, El-Moussely LM, Ismail DK. Pseudomonas aeruginosa biofilm formation and quorum sensing lasR gene in patients with wound infection. Egypt J Med Microbiol. 2016;25:101–108. doi: 10.12816/0037098. [DOI] [Google Scholar]
- 32.Li H, Li X, Wang Z, Fu Y, Ai Q, et al. Autoinducer-2 regulates Pseudomonas aeruginosa PAO1 biofilm formation and virulence production in a dose-dependent manner. BMC Microbiol. 2015;15:192–200. doi: 10.1186/s12866-015-0529-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J Bacteriol. 2005;187:3898–3902. doi: 10.1128/JB.187.11.3898-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa . Protein Cell. 2015;6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aboushleib HM, Omar HM, Abozahra R, Elsheredy A, Baraka K. Correlation of quorum sensing and virulence factors in Pseudomonas aeruginosa isolates in Egypt. J Infect Dev Ctries. 2015;9:1091–1099. doi: 10.3855/jidc.6492. [DOI] [PubMed] [Google Scholar]
- 36.Henkel M, Schmidberger A, Kühnert C, Beuker J, Bernard T, et al. Kinetic modeling of the time course of N-butyryl-homoserine lactone concentration during batch cultivations of Pseudomonas aeruginosa PAO1. Appl Microbiol Biotechnol. 2013;97:7607–7616. doi: 10.1007/s00253-013-5024-5. [DOI] [PubMed] [Google Scholar]
- 37.Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reuter K, Steinbach A, Helms V. Interfering with bacterial quorum sensing. Perspect Medicin Chem. 2016;8:PMC-S13209. doi: 10.4137/PMC.S13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerra-Santos L, Käppeli O, Fiechter A. Dependence of Pseudomonas aeruginosa continous culture biosurfactant production on nutritional and environmental factors. Appl Microbiol Biotechnol. 1986;24:443–448. doi: 10.1007/BF00250320. [DOI] [Google Scholar]
- 40.Schaber JA, Carty NL, McDonald NA, Graham ED, Cheluvappa R, et al. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2004;53:841–853. doi: 10.1099/jmm.0.45617-0. [DOI] [PubMed] [Google Scholar]
- 41.Karatuna O, Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin Microbiol Infect. 2010;16:1770–1775. doi: 10.1111/j.1469-0691.2010.03177.x. [DOI] [PubMed] [Google Scholar]