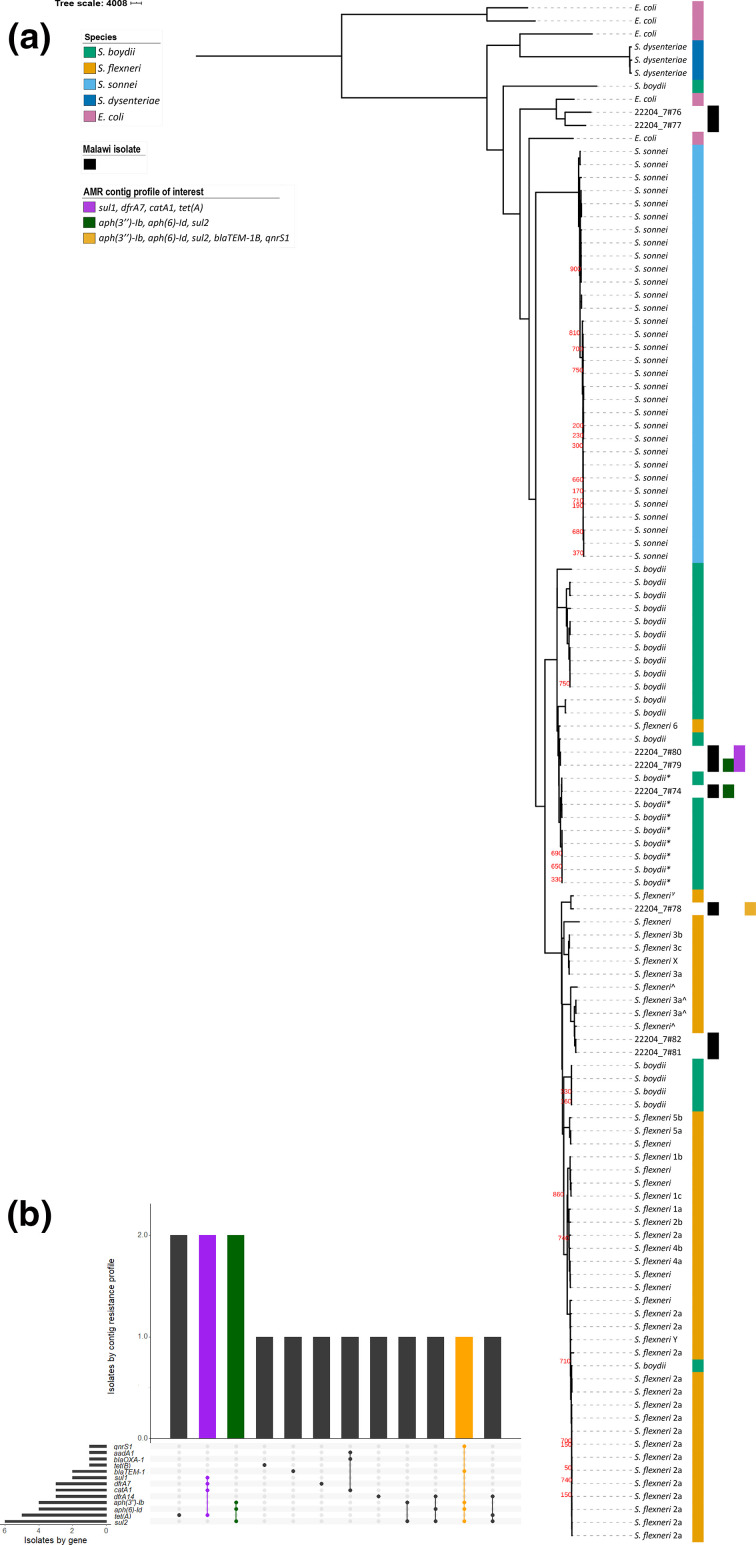
Fig. 1.
Maximum-likelihood phylogeny of eight isolates from Malawi, contextualized among Escherichia coli and Shigella and highlighting some AMR profiles of interest. AMR profiles of interest are those of contiguous sequences which are multi-drug-resistant and shared across multiple isolates, and in one isolate carries additional AMR genes including an FQR gene (qnrS1). They are indicated by columns to the right of the tree (a) and by coloured bars in the AMR profile chart (b). (a) Columns to the right of the phylogeny indicate, from left to right, species, study isolate and AMR profiles of interest. GTR+G substitution model, 1000 bootstrap validation and mid-point rooted. * S. boydii clade 3, ^ S. flexneri phylogroup 2, γ S. flexneri phylogroup 7. All bootstrap values for internal nodes with support <900 are displayed. Bar, SNPs per site. (b) Intersection of individual AMR genes by isolate and AMR gene profile by contig.