FIGURE 1.
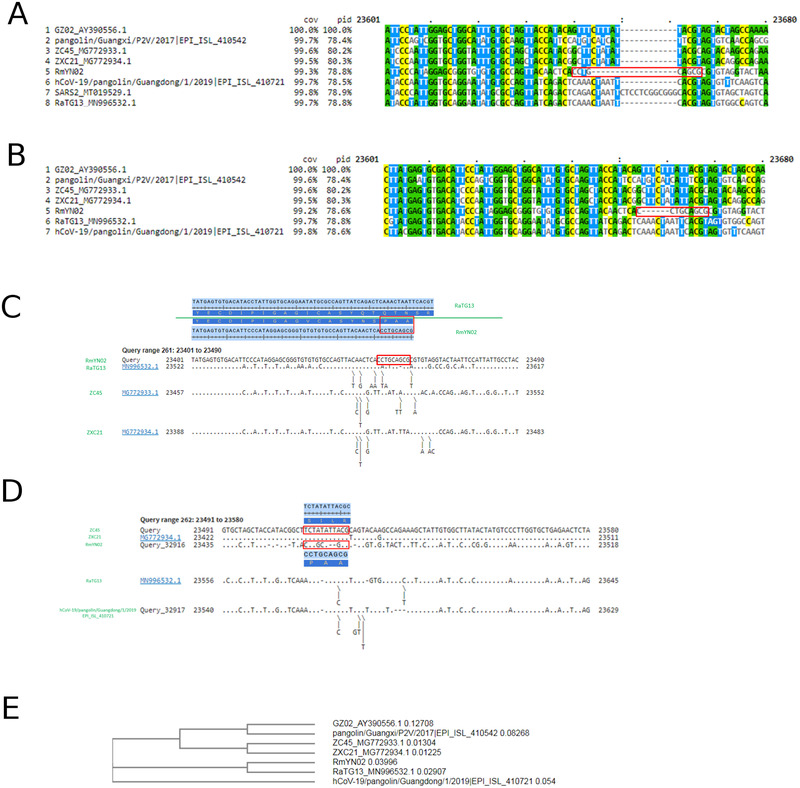
(A) Clustal W multiple sequence alignment of RmYN02 with the strains used in Zhou et al. for comparison. RmYN02's nucleotides coding the PAA amino acids (CCT GCA GCG) are surrounded by a red box. No insertion in RmYN02 is visible; on the contrary, a deletion splitting the nucleotides coding for PAA is observed. (B) Clustal W multiple sequence alignment of RmYN02 with the strains used in Zhou et al. for comparison, with the exception of SARS‐CoV‐2. The deletion characterizing RmYN02 at the S1/S2 junction appears to cause a split of the first nucleotide from the rest of the sequence coding for the PAA amino acids (CCT GCA GCG, surrounded by a red box). (C) Pairwise comparisons of RmYN02 (anchor) to RaTG13, ZC45, and ZXC21. No PAA insertion is observed in RmYN02 in these comparisons. (D) Pairwise comparisons of ZC45 (anchor) to ZXC21, RmYN02, RaTG13, and Pangolin/GD/2019. RmYN02's nucleotides coding the PAA amino acids (surrounded by a red box) are aligned as mutations relative to ZC45 rather than insertions. (E) Phylogenetic tree of SARS‐GZ02, Pangolin/GX/2017, ZC45, ZXC21, RmYN02, RaTG13, and Pangolin/GD/2019 produced by CLUSTAL W based on the alignment of their genomes as in (B)
