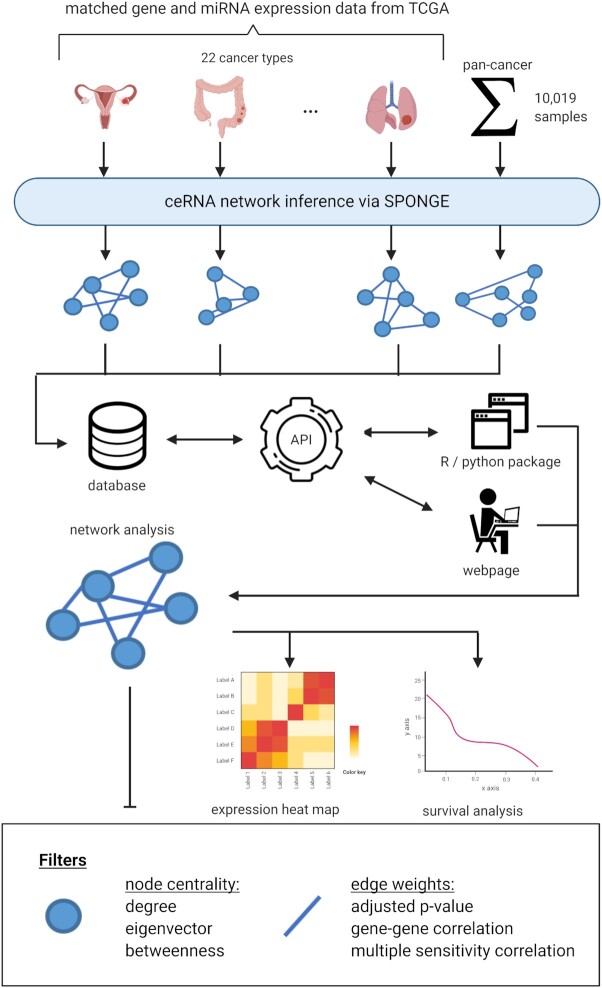
Figure 1.
SPONGEdb overview: ceRNA networks for 22 cancer types as well as a pan-cancer ceRNA network for all 10 019 samples in TCGA have been inferred with SPONGE (26) and can be queried efficiently via a RESTful API through any programming language. We provide R and Python packages to simplify scripted access and offer a user-friendly and feature-rich web interface offering additional insights into ceRNA from external resources (ENCODE (34) for additional gene information, miRBase for additional miRNA information, Quick GO for correlating Gene Ontologies and the WikiPathways keys for each gene). The web interface offers visualization of the ceRNA networks, expression heat maps and survival analysis via Kaplan–Meier plots. Various filters for node centrality and edge weights allow refining the ceRNA network to focus on cancer-type-specific essential ceRNA regulators and their strongest or most significant interactions.