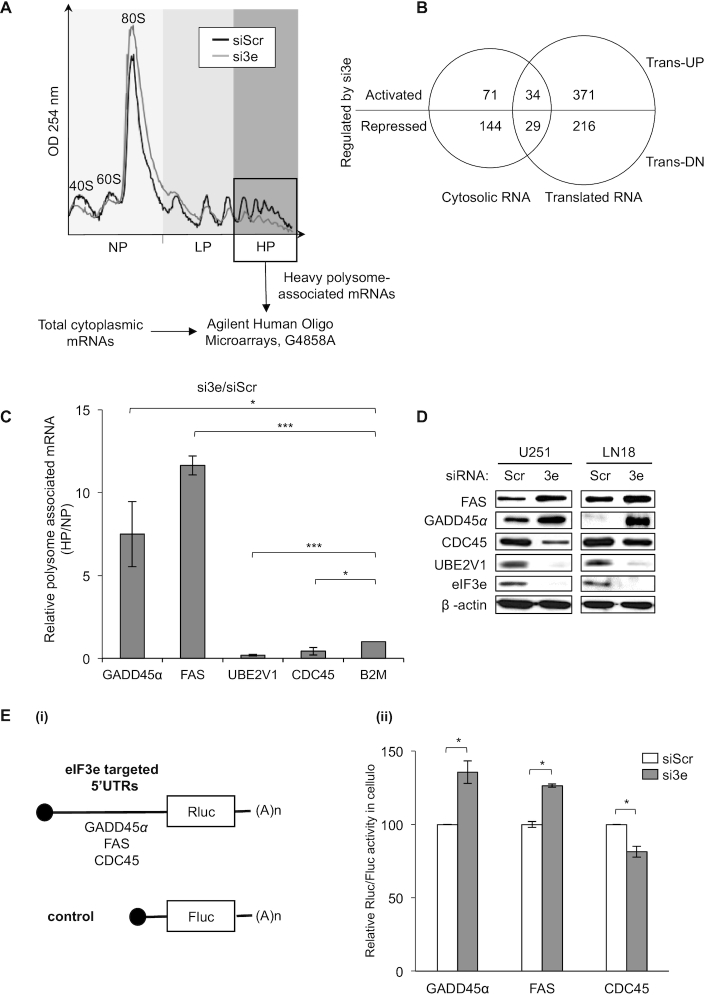
Figure 2.
eIF3e drives selective mRNA translation of mRNAs involved in GBM tumor progression and response to treatments. (A) Polysome profile of U251 cells treated with control (siScr) or eIF3e (si3e) siRNAs. mRNA was isolated from fractions containing three ribosomes (box) and analyzed in parallel with cytoplasmic mRNA using microarrays (see the ‘Materials and Methods’ section). The peaks corresponding to the 40S, 60S and 80S ribosomal subunits and polysomes are indicated, as well as NP, LP and HP fractions. (B) Microarray data analysis using a Venn diagram showing the overlap between cytosolic mRNAs and translationally regulated mRNAs. The number of eIF3e activated or repressed mRNAs in each group is indicated. (C) Validation of the genome-wide polysome profiling in (A) by RT-qPCR analysis from pooled NP and HP fractions using specific primers for the indicated mRNAs and quantification by analyzing the ratio HP/NP [error bars ± SEM of three experiments; *P < 0.05, ***P < 0.0005 (paired t-test)]. (D) Western blot analysis of the indicated proteins in U251 and LN18 cells treated with control (siScr) or eIF3e (si3e) siRNAs. (E) Ratio of Renilla/Firefly luciferase activities (Rluc/Fluc) (ii) determined using U251 cells treated with control (siScr) and eIF3e (si3e) siRNAs, followed by co-transfection with Rluc and Fluc mRNA reporters depicted in (i) [error bars ± SEM of three independent experiments; *P < 0.05 (paired t-test)].