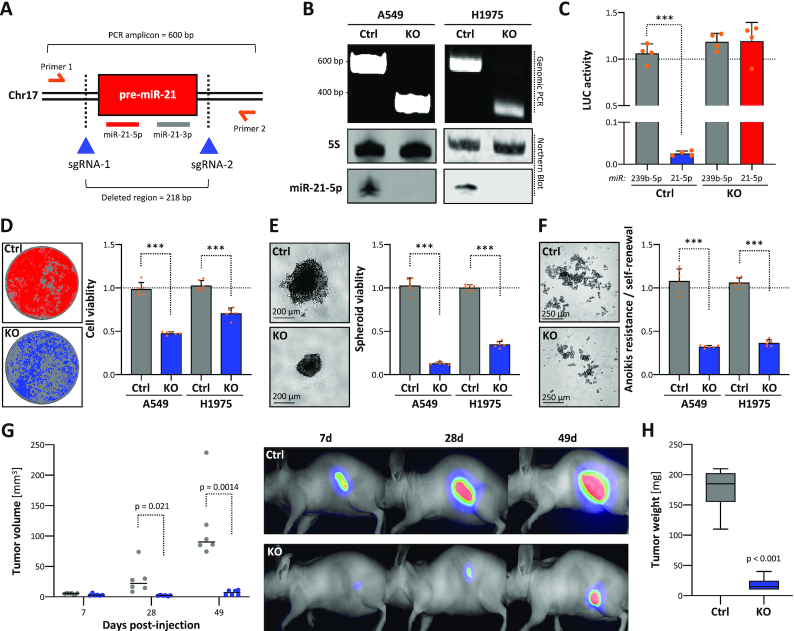
Figure 2.
Genomic deletion of MIR21 in cancer impairs tumor growth. (A) Schematic showing the experimental strategy to delete the MIR21 locus by using Cas9 nuclease and indicated CRISPR guide RNAs (sgRNAs). Primers for PCR on genomic DNA (gDNA) and amplicon/deleted region sizes are indicated. (B) Representative PCR analysis on gDNA of parental (Ctrl) and MIR21-deleted (KO) A549 and H1975 cells (top panel). Representative northern blot analysis of miR-21-5p expression in Ctrl or KO A549 and H1975 cells (bottom panel). 5S served as a normalization control. (C) miRNA reporter analyses in A549 Ctrl or miR-21 KO cells. The activity of indicated miRNAs (cel-miR-239b-5p and hsa-miR-21-5p) was analyzed by using antisense luciferase reporters. Luciferase activities, normalized to a control reporter comprising a minimal 3′-UTR, were determined in four experiments. Viability in 2D (D) or 3D spheroid growth (E) and anoikis resistance (F) analyses of parental (Ctrl) or miR-21 KO A549 and H1975 cells. Representative images of A549 cells are shown (left panels). Viability and anoikis resistance were determined by CellTiter Glo in six median-normalized experiments (right panels). (G) Ctrl and miR-21 KO A549 cells expressing iRFP were injected (s.c.) into nude mice (n = 6 per condition). Xenograft tumor volumes were measured (left panel) and monitored by near-infrared imaging (right panel) at indicated time points upon injection. (H) Box plots showing final tumor mass 49 days post-injection. Statistical significance, indicated by P-values, was determined by the Mann–Whitney test: ***P < 0.001.