Abstract
Gene correction is a valuable strategy for treating inherited retinal degenerative diseases, a major cause of irreversible blindness worldwide. Single gene defects cause the majority of these retinal dystrophies. Gene augmentation holds great promise if delivered early in the course of the disease, however, many patients carry mutations in genes too large to be packaged into adeno-associated viral vectors and some, when overexpressed via heterologous promoters, induce retinal toxicity. In addition to the aforementioned challenges, some patients have sustained significant photoreceptor cell loss at the time of diagnosis, rendering gene replacement therapy insufficient to treat the disease. These patients will require cell replacement to restore useful vision. Fortunately, the advent of induced pluripotent stem cell and CRISPR-Cas9 gene editing technologies affords researchers and clinicians a powerful means by which to develop strategies to treat patients with inherited retinal dystrophies. In this review we will discuss the current developments in CRISPR-Cas9 gene editing in vivo in animal models and in vitro in patient-derived cells to study and treat inherited retinal degenerative diseases.
Keywords: genome engineering, CRISPR-Cas9, induced pluripotent stem cells, retinal degeneration, transplantation
1. Introduction
Inherited retinal degenerative disorders, which are predominantly caused by single gene defects, are a major cause of irreversible blindness worldwide. As evident from the numerous trials of adeno-associated viral vector (AAV) gene augmentation published to date (Bainbridge et al., 2015; 2008; Cideciyan et al., 2009; Hauswirth et al., 2008; Jacobson et al., 2012; Maguire et al., 2009; 2008; Russell et al., 2017; Simonelli et al., 2010; Testa et al., 2013; Weleber et al., 2016), gene therapy delivered early in the course of disease holds great promise, especially for recessive conditions. Unfortunately, many patients have disease-causing mutations in genes that are too large to be packaged into AAVs (Dong et al., 1996; Wu et al., 2010). For instance, gene therapy for mutations in the genes ABCA4 and USH2A, which together account for almost 25% of inherited retinal disease (Stone et al., 2017), requires the delivery of cDNAs that are two to four times larger than the AAV packaging capacity. Thus, a substantial fraction of degenerative retinal diseases are not amenable to gene augmentation therapy with current viral vectors. Moreover, the retina is highly sensitive to transgene expression levels and for many genes overexpression from strong, heterologous promoters leads to cytotoxicity (Burnight et al., 2014; Luo et al., 2011; Olsson et al., 1992; Seo et al., 2013; Tan et al., 2001). For patients who have sustained significant photoreceptor cell loss, gene therapy will not be sufficient and some form of photoreceptor cell replacement will be required to restore useful vision. The advent of induced pluripotent stem cell (iPSC) (Gu et al., 2015; Park et al., 2008; Takahashi and Yamanaka, 2006; J. Yu et al., 2007) affords researchers and clinicians the ability to generate therapeutic cells from the patients for whom they are intended. Unlike genetically complex diseases such as age related macular degeneration, the treatment of Mendelian disorders such as retinitis pigmentosa (RP), with cells derived from autologous iPSCs, will likely require correction of the patient’s disease-causing gene prior to cellular differentiation and transplantation. As indicated above, many retinal genes have very large coding sequences and require exquisite transcriptional control. For these reasons, it would be very valuable to have some means of editing genes in vivo early in disease progression prior to significant photoreceptor cell death and in vitro late in disease progression when photoreceptor cell replacement is required.
The recent discovery that prokaryotic immune components known as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated nucleases such as Cas9 can mediate genome editing in mammalian cells provides a means to correct disease-causing mutations while leaving the gene under control of its endogenous regulatory elements (Jinek et al., 2013a; Mali et al., 2013a; Cong et al., 2013). The powerful combination of this new technology with induced pluripotent stem cells provides researchers with the ability to develop treatments for inherited retinal degenerative blindness regardless of disease state. In this review, we discuss CRISPR-Cas9 gene editing, patient-specific iPSCs and the therapeutic potential afforded by combining the two technologies to study and treat inherited retinal disease.
2. The CRISPR system
2.1. Discovery and role in bacteria
With the recent explosion of CRISPR based genome editing technologies, it might be somewhat surprising to learn that CRISPRs were first observed in strains of bacteria in the 1980s. Originally referred to as Short Regularly Spaced Repeats (SRSRs) before the CRISPR acronym was adapted, the first CRISPR array was discovered in E. coli and contained 14 direct repeats consisting of highly homologous sequences of 29 nucleotides separated by 32 nucleotides that served as spacers between each repeat (Ishino et al., 1987). Soon after, arrays of tandem repeats were discovered in additional strains of bacteria and various archaeal lineages, including extremophilic organisms. As of 2013, Barrangou and van der Oost reported that CRISPR arrays have been detected in more than 85% of sequenced archaeal genomes and 49% of sequenced bacterial genomes (Barrangou and van der Oost, 2015).
The presence of CRISPR arrays within intergenic regions of the genome led some to hypothesize that they may help to modulate gene transcription (Hermans et al., 1991; Nakata et al., 1989). It wasn’t until 2005, however, that CRISPRs were first linked to invading genetic elements (Mojica et al., 2005; Pourcel et al., 2005). In these studies, it was noted that: 1) BLAST searches of sequenced bacterial species revealed that ~2% of analyzed spacers within CRISPR arrays showed striking similarity to sequences from viral DNAs, 2) that these sequence matches were generally observed in genetic elements that may be capable of invading hosts harboring said CRISPR arrays with matching spacer regions and 3) that CRISPR arrays may be able to acquire new spacers from foreign invading elements (Mojica et al., 2005; Pourcel et al., 2005). These findings lead to the hypothesis that CRISPR arrays may be a newly-discovered adaptable immune defense system against invading genetic elements (Mojica et al., 2005; Pourcel et al., 2005). The observation that the larger the CRISPR array in bacteria, the fewer the number of phages capable of successfully infecting them supports this immune defense hypothesis (Bolotin et al., 2005).
2.2. Components and classification of CRISPR-Cas systems
Although CRISPR-Cas systems are classified into three distinct types (Class I, II or III) (Bhaya et al., 2011; Makarova et al., 2011; Wiedenheft et al., 2012), they are composed of the same basic components. Generally, CRISPR-Cas systems consist of one or more arrays of alternating repeat sequences and spacers, a leader sequence and a set of CRISPR-associated (cas) genes. Cas genes produce CRISPR-RNAs (crRNAs) and Cas proteins which, as summarized in Figure 1, function in several ways following infection by a foreign invader (1): uptake of new spacers from foreign DNA elements (2; acquisition), generation of processed smaller crRNAs from CRISPR transcripts (3; expression), and targeting and cleavage of invading genetic elements (4; interference) via protein-crRNA complexes that bind with complementary spacer sequences (Leenay and Beisel, 2017).
Figure 1. The CRISPR-Cas systems function in bacteria and archea as adaptive immune systems against foreign genetic material.
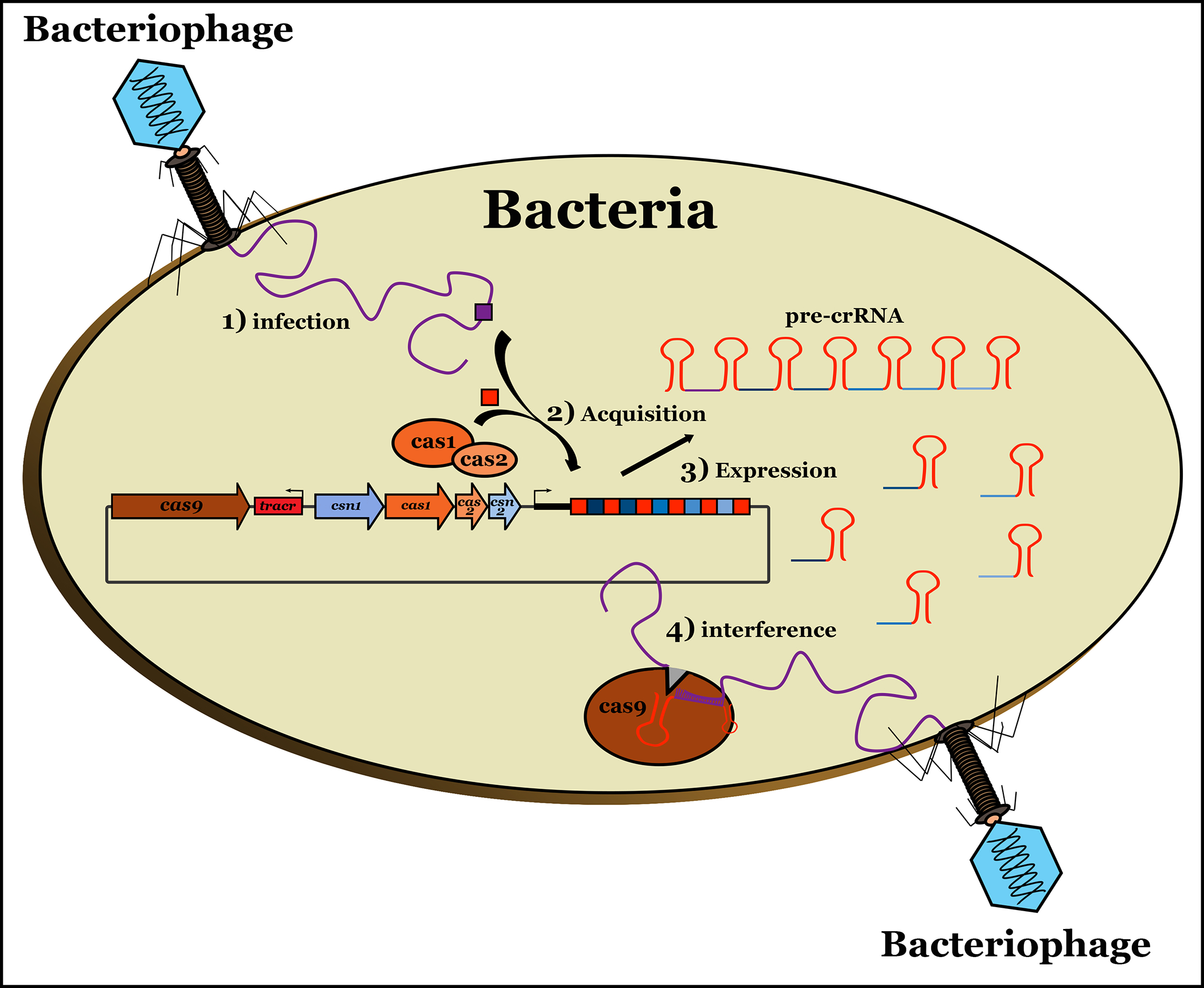
The system is composed of a CRISPR array of alternating conserved repeats and target-specific spacers (protospacers) acquired from fragments of foreign genetic material. The bacterium creates a heritable memory of infection. Upon entry of a foreign invader (1 – infection), foreign DNA sequences are incorporated into the bacterial CRISPR locus (2 – acquisition) and subsequently the bacterium transcribes CRISPR RNAs (crRNAs) from the array which associate with Cas effector proteins to create a ribonucleoprotein surveillance complex (RSC) (3 – expression). The RSC recognizes a sequence directly downstream of the crRNA target sequence – the protospacer adjacent motif (PAM). Following guide binding, the Cas nuclease cleaves the target DNA leading to the clearance of the foreign invader (4 – interference) (Leenay and Beisel, 2017).
The three classes of CRISPR-Cas systems are characterized by the distinct sets of cas genes that each express. Each system utilizes two universal cas genes, cas1 and cas2 whose main function is in spacer acquisition (Marraffini and Sontheimer, 2009; Beloglazova et al., 2008). Beyond these two genes each class is characterized by a unique cas gene. Type I CRISPR-Cas systems encode the gene cas3, which functions as an ATP-dependent helicase and a single-stranded DNA nuclease to assist in small guide crRNA-mediated interference of foreign DNA elements (Sinkunas et al., 2011). Type II CRISPR-Cas systems include the gene, cas9, which produces a large protein capable of generating crRNAs and cleavage of target DNA through two nuclease domains, a RuvC-like domain near the amino terminus and an HNH nuclease domain located centrally (Makarova et al., 2011). Finally, Type III CRISPR-Cas systems express cas10, which produces a protein that contains a domain homologous to the palm domain of nucleic acid polymerases and nucleotide cyclases (Barrangou and van der Oost, 2013; Makarova et al., 2011; Cocozaki et al., 2012). While each class of CRISPR-Cas plays a vital role in bacteria and archaeal viral immunity, as discussed below, the Type II CRISPR system and, in particular the Cas9 protein, has recently dominated the field of genome editing. For an in-depth review of the origin and history of CRISPR-Cas systems in immunity, the reader is encouraged to refer to elegant reviews (Barrangou and van der Oost, 2013; Hsu et al., 2014; Sander and Joung, 2014).
2.3. Early CRISPR-Cas-mediated genome editing studies in eukaryotic cells
In 2013 the field of genome editing was transformed by the publication of three seminal papers that described, for the first time, the use of components of the type II prokaryotic CRISPR adaptive immune system to perform targeted genomic modification in eukaryotic cells (Jinek et al., 2013a; Mali et al., 2013a; Cong et al., 2013). Although genome editing technologies such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) demonstrated the ability to execute site-specific cleavage of eukaryotic DNA, the use of ZFNs has been limited by the need to engineer a specific protein for each specific DNA target site, which is time consuming and expensive; likewise, TALENs require a 5’ thymidine residue which may limit target site availability (Bogdanove and Voytas, 2011; Urnov et al., 2010). The CRISPR-Cas-mediated approach offered a new means for targeted genome modification that was scalable, relatively low-cost and easier to engineer.
Between 2011–2012, multiple findings laid the groundwork for the potential use of CRISPR-Cas components in eukaryotic cells. The Type II CRISPR-Cas systems require only a single protein to cleave DNA, Cas9 (Sapranauskas et al., 2011). Cas9 accomplishes target cleavage through interaction with a 20 nucleotide guide sequence contained within an associated crRNA transcript and a partially-complimentary trans-activating crRNA (tracrRNA). This crRNA:tracrRNA complex can be redesigned as a single guide RNA (sgRNA) that could supply the needed site recognition and binding information required for Cas9. Target cleavage also requires recognition of a sequence on the opposite strand of guide pairing called the protospacer adjacent motif (PAM) (Jinek et al., 2012).
In eukaryotic cells, using a human-codon-optimized version of the Streptococcus pyogenes Cas9 (SpCas9) engineered with a nuclear localization signal and an sgRNA targeting the human clathrin light chain 1 (CTLA1) gene, they demonstrated site-specific double-strand breaks (DSBs) in HEK293T cells (Jinek et al., 2013a). Additional experiments using extracts from transfected HEK293T cells suggested that the limiting factor for Cas9 function is presence of sgRNA and/or its loading into Cas9 (Jinek et al., 2013a). To increase assembly of sgRNA and Cas9, these investigators tested the effect of extending the Cas9-binding region of the guide RNA by engineering two additional versions of the CLTA1 sgRNA: one that included an additional four or ten base pairs within the helix where interactions between the crRNA and tracrRNA occur and another in which the 3’-end of the sgRNA was extended by five nucleotides based on the native sequence of the S. pyogenes tracrRNA. Each new CLTA1 sgRNA produced more efficient Cas9 function in HEK293T cells than the original (Jinek et al., 2013a).
Cas9-mediated induction of DSBs often leads to DNA repair via the error-prone mechanism of non-homologous end-joining (NHEJ) (Mali et al., 2013a; Cong et al., 2013). Conversion to a DNA nickase by engineering an aspartate-to-alanine substitution (D10A) within the RuvC I domain of the SpCas9 protein improves specificity in mammalian cells (Mali et al., 2013a; Cong et al., 2013). Nicked DNA is typically repaired precisely via single-stranded base repair (SSBR) mechanisms (Davis and Maizels, 2014) or by homology-directed repair (HDR). Using this approach along with co-delivery of a DNA homology repair template, researchers can introduce targeted genomic insertions (Mali et al., 2013a; Cong et al., 2013). Together, these innovative studies paved the way for a new field of genome editing; which when combined with the use of patient-specific induced pluripotent stem cells, have great implications for translational research.
2.4. Improving CRISPR-Cas for greater specificity
2.4.1. Orthologs of Streptococcus pyogenes Cas9
Given the initial success of CRISPR-Cas9-mediated genome editing in human cells, the natural progression of investigation for many scientists, particularly with respect to clinical translatability, was a move towards genome editing in vivo. The potential advantages of correcting a mutation in the affected cells, while leaving the gene’s regulatory sequences completely intact, are enormous. However, in addition to requiring exquisite precision, for CRISPR based genome editing to be useful in vivo, an efficient delivery system would likely be needed. One potential avenue would be to use adeno-associated viral vectors, which have been successfully employed for gene augmentation based treatment of a variety of disorders (Ghazi et al., 2016; H. Jiang et al., 2006; Kay et al., 2000; Maguire et al., 2008; Mendell et al., 2015; Mueller et al., 2017; Smith et al., 2017; Wagner et al., 1998; Worgall et al., 2008). The carrying capacity of AAV is limited to approximately 4.9 kilobases (Dong et al., 1996). Cargo of up to 5.2 kb in length have been packaged, however, the packaging efficiency decreases precipitously with insert size (Grieger and Samulski, 2005). Packaging SpCas9 (4.2 kilobases) and a sgRNA (under control of a promoter) in the same vector would be inefficient and studies to date have employed dual vector AAV-CRISPR-SpCas9 systems to achieve in vivo delivery (Xionggao Huang et al., 2017; Hung et al., 2016; Ruan et al., 2017; Swiech et al., 2015). To this end, investigators employed smaller, alternative CRISPR effector proteins orthologous to SpCas9 (E. Kim et al., 2017; Ran et al., 2015), several of which are discussed in Table 1 below.
Table 1.
Cas9 orthologs
| Species | Size (aa) | PAM | Target (nt) | Reference |
|---|---|---|---|---|
| Streptococcus pyogenes Cas9 | 1368–1424 | NGG | 20 | (Jinek et al., 2012) |
| Staphylococcus aureus Cas9 | 1053 | NNGRRT | 20–24 | (Kleinstiver et al., 2015) |
| Streptococcus thermophiles 1 Cas9 | 1122 | NNNRRT | 21 | (Horvath et al., 2008) |
| Streptococcus thermophiles 3 Cas9 | 1393 | NGGNG | 19 | (Horvath et al., 2008) |
| Neisseria meningitidis Cas9 | 1109 | NNNNGATT | 23–24 | (Hou et al., 2013) |
| Francisella novicida Cas9 | 1629 | NGG | 22 | (H. Hirano et al., 2016) |
| Treponema denticola Cas9 | 1423 | NAAAAN | 20 | (Anders et al., 2014) |
| Acidaminococcus Cas12a (Cpf1) | 1308–1310 | TTTV | 23–24 | (Zetsche et al., 2015) |
| Lachnospiraceae Cas12a (Cpf1) | 1228 | TTTV | 23–24 | (Zetsche et al., 2015) |
| Leptotrichia buccalis Cas13a (C2c2) | 1159 | none | 20–28 | (Liu et al., 2017) |
| Leptotrichia shahii Cas13a (C2c2) | 1389 | H | 14–28 | (Abudayyeh et al., 2016) |
| Campylobacter jejuni Cas9 | 984 | NNNNACAC or NNNRYAC | 22 | (E. Kim et al., 2017) |
Staphylococcus aureus Cas9
Staphylococcus aureus Cas9 (SaCas9) targets sequences between 21 and 24 base pairs in length, but also requires a six nucleotide protospacer adjacent motif (PAM site) consisting of 3’ NNGRRT directly downstream of the guide sequence (where N indicates any nucleotide and R indicates the purine nucleotides A or G). SaCas9 has displayed similar gene targeting efficiencies and increased specificity when compared side-by-side with SpCas9 (Ran et al., 2015). The requirement for a longer PAM sequence, which is likely to account for its increased specificity, results in fewer compatible putative target sites throughout the genome, which could limit its utility (Cebrian-Serrano and Davies, 2017). However, the shorter size of SaCas9 cDNA (~1 kilobase) makes it more amenable for packaging into AAV and has been shown to mediate editing in vivo in both mice (Ran et al., 2015) and pigs (Burnight et al., 2017) and ex vivo in hematopoietic stem cells (Ye et al., 2016) via AAV-mediated delivery.
Cas12a (Cpf1)
Cpf1, more recently re-named Cas12a (Shmakov et al., 2015), is another Type II CRISPR effector protein in bacteria that has three key differences from other Cas9 systems; 1) Cas12a is structurally distinct from Cas9 proteins and can bind target DNA through a Cas12a:crRNA complex and does not require a tracrRNA (Zetsche et al., 2015); 2) Cas12a:crRNA complexes recognize a 5’ T-rich (TTTV where V indicates A, C, or G nucleotides) PAM site upstream of the target sequence, unlike the 3’ G-rich PAM sites utilized by Cas9 systems that lie downstream of the target sequence (Zetsche et al., 2015); and 3) as opposed to Cas9-generated blunt ends, Cas12a cleaves target DNA via a staggered DSB leaving a 4 or 5 nucleotide 5’ overhang (Fonfara et al., 2016; Zetsche et al., 2015), which may allow for directional insertion of donor DNA into the genome via the more efficient NHEJ than HDR-mediated mechanisms that are particularly challenging in terminally differentiated, non-dividing cells (Zetsche et al., 2015).
Streptococcus thermophilus Cas9
To date, two Streptococcus thermophiles Cas9s (StCas9s) have been modified and tested for genomic modification in mammalian cells: St1Cas9 from the CRISPR1 locus (Cong et al., 2013) and St3Cas9 from the CRISPR3 locus (Xu et al., 2015). Side-by-side comparison of St1Cas9 with SpCas9 and SaCas9 showed successful modification in human cells, but a lower level of overall activity (Ran et al., 2015). Experiments comparing St1Cas9, St3Cas9 and SpCas9 showed robust cleavage efficiencies for each in human cells and that off-target mutagenesis was lower in cells treated with either St1Cas9 or St3Cas9 compared to SpCas9 (Müller et al., 2016). Similar to observations in cells treated with SaCas9, this increased specificity may be due to the need for more complex PAM sites for St1Cas9 (NNAGAAW where W indicates A or T nucleotides) and St3Cas9 (NGGNG).
Additional Cas9 orthologs
In addition to those described above, Cas9 orthologs have been identified in Neisseria meningitidis (NmCas) (Q. Zhang et al., 2013), Francisella novicida (FnCas9) (Sampson and Weiss, 2013) and Campylobacter jejuni (CjCas9) (E. Kim et al., 2017). Like SaCas9, NmCas9 recognizes a more stringent PAM sequence (NNNNGATT), is smaller than SpCas9 (Cebrian-Serrano and Davies, 2017) and has been shown to induce HDR in human cells (Esvelt et al., 2013; Hou et al., 2013). Likewise, CjCas9 recognizes more complex PAM sequences (NNNNACAC or NNNNRYAC) and is more specific than SpCas9 and SaCas9 (E. Kim et al., 2017). FnCas9 is the largest of the Cas9 orthologs described to date; it has not been shown to induce modification in mammalian cells, however was successful following microinjection of mouse zygotes (H. Hirano et al., 2016). For a more extensive analysis of Cas9 orthologs, please see the comprehensive review by Cebrian-Serrano and Davies (Cebrian-Serrano and Davies, 2017).
2.5. Biological applications of the CRISPR-Cas system
As indicated above, since discovering that the CRISPR-Cas system could be used to modify the genome of mammalian cells, a myriad of genome editing experiments have been performed. Numerous studies have now described how the CRISPR-Cas system can be used to 1) delete an existing gene (Buchrieser et al., 2017; P. Wang et al., 2015), 2) drive expression of a mutant gene product (Yumlu et al., 2017; J.-P. Zhang et al., 2017), or 3) increase or decrease host gene expression (Heman-Ackah et al., 2016).
2.5.1. Targeted gene correction
One of the most widely used applications of the CRISPR-Cas technology has been targeted gene correction. Depending on the mutation, CRISPR-Cas-induced DSBs are predominantly repaired via either NHEJ or homology-dependent repair. If the mutation occurs deep within intronic sequence, a single guide or pair of guides can direct Cas mediated cleavage near or surrounding the mutation which, via NHEJ, can remove the mutant sequence and restore normal gene function (Burnight et al., 2017; Iyombe-Engembe et al., 2016; Ouellet et al., 2017). This process is efficient in that it does not rely on the rate-limiting step of homology-dependent repair. Additionally, employing Cas-induced NHEJ to disrupt the reading frame of a gene or allele that carries a dominant-negative gain-of-function mutation can potentially prevent mutation induced disease progression (Bakondi et al., 2016; Burnight et al., 2017; Monteys et al., 2017; Shin et al., 2016; Yamamoto et al., 2017). However, if the mutation lies close to or within exonic space (i.e., sequence that includes a splice donor or acceptor site), homologous dependent repair, which relies on co-delivery of CRISPR-Cas with wild-type (i.e., unmutated DNA sequence) homologous donor sequence, will likely be needed (Burnight et al., 2017; Xiaosong Huang et al., 2015; J.-P. Zhang et al., 2017).
2.5.2. Regulation of gene expression
To modulate levels of gene expression, various groups have taken advantage of knowledge gained from elucidation of the crystal structure of S. pyogenes Cas9, which revealed two nuclease domains, RuvC and HNH, that cleave the non-complementary and complementary target DNA strands, respectively. Mutating a single amino acid in each of the RuvC (D10A) and HNH (H840A) domains creates a catalytically inactive Cas9 (hereafter referred to as dCas9) (Anders et al., 2014; Jinek et al., 2014; Nishimasu et al., 2014). The dCas9 protein lacks the exonuclease function of the Cas protein while retaining its binding properties to DNA-RNA hybrids. When fused to a transcriptional activator domain and co-expressed with a guide or guides, the CRISPR-dCas9 recruits transcriptional agonists to promoters to increase expression of target genes (CRISPRa). Alternatively, dCas9-repressor fusions interfere with transcriptional elongation, RNA polymerase or transcription factor binding (CRISPRi) (Cheng et al., 2013; Gilbert et al., 2013; Kearns et al., 2014; Maeder et al., 2013), which could be used to knock down gene expression. For instance, when fused to a human codon-optimized dCas9 from S. pyogenes, the repressive chromatin modifier domains Kruppel-associated box (KRAB), chromo shadow (CS) or the Trp-Arg-Pro-Trp (WRPW) motif specifically represses target protein expression (Gilbert et al., 2013).
Similar gene repression strategies have been reported using dCas9-VPR (Heman-Ackah et al., 2016), doxycycline-inducible dCas9 fused to a KRAB repression domain (Mandegar et al., 2016), CRISPRi and CRISPRa systems (Mandegar et al., 2016). Repression has been reported to be most effective when sgRNAs are targeted to enhancers, proximal promoters, and the coding region downstream from the transcription start site (Du and Qi, 2016). These strategies may be useful for modulating disease progression in dominantly inherited forms of disease such as retinitis pigmentosa (RP). For instance, the single nucleotide polymorphism rs7984 occurs 93 bp upstream of the Pro23His mutation in the RHO gene (Burnight et al., 2017), one of the most common disease-causing variants in Rhodopsin-associated retinal degeneration (Hartong et al., 2009; Stone et al., 2017). This mutation occurs near a PAM site and is only 69 bp downstream of the TSS and thus could be used to selectively-target and knockdown expression of the disease-causing allele. Specific application of this technology to retinal diseases is discussed in section 4 below.
2.6. Specificity of CRISPR-Cas9-mediated editing in mammalian cells
CRISPR-Cas9 holds great potential for treating inherited eye disease; however development of precise reagents that specifically target the desired locus while inducing minimal off target modifications is an important step toward moving genome editing into the clinic. The generation of Cas9 variants with increased specificity addresses off-targeting issues. As indicated above, when combined with paired, offset sgRNAs complementary to opposite strands of the target site, a Cas9 mutant carrying a D10A variation in the RuvC nuclease domain of S. pyogenes Cas9 nicks both strands of DNA in a manner similar to that of dimeric zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) (Gasiunas et al., 2012; Jinek et al., 2012; Cong et al., 2013). This paired nicking strategy reduced off-target activity by 50–1000 fold in human cells (Ran et al., 2013a). Structure-guided protein engineering improves the specificity of SpCas9 (Kleinstiver et al., 2016; Slaymaker et al., 2016; Chen et al., 2017). By interfereing with the target DNA-Cas9 contacts via amino acid substitutions, engineered Cas9 variants significantly reduce off-target mutations and increase target locus specificity. Disruption of the connections at N497, R661, Q695, and Q926 (referred to as SpCas9-HF1) relaxed the non-specific interaction between target DNA and SpCas9 (Kleinstiver et al., 2016). Two mutants, 1) one carrying a single alanine substitution [SpCas9 (K855A)] and 2) one carrying three alanine substitutions [eSpCas9(1.1); K848A/K1003A/K1060A] were evaluated for genome-wide specificity using BLESS (discussed further below in section 2.7.1.). Both mutants significantly reduced off-target indel formation compared to WT SpCas9. Moreover, no new off-target sites were generated (Slaymaker et al., 2016).
Evaluation of the activity of SpCas9-HF and eSpCas9(1.1) mutants with alanine substitutions at conserved residues near the RNA-DNA interface revealed that both eSpCas9-HF1 and epSpCas9(1.1) are trapped in an inactive state when bound to mismatched targets (Chen et al., 2017). sgRNAs with single mismatches against the human FANCF gene and a quadruple mutant (N692A/M694A/Q695A/H698A, referred to as HypaCas9) demonstrated greatly reduced cleavage activity at each sgRNA nucleotide compared to WT SpCas9 and both SpCas9-HF1 and eSpCas9(1.1). Off-target cleavage from three sgRNAs previously shown to exhibit substantial off-target activity and three previously uncharacterized sgRNAs with a moderate number of in silico sites predicted was evaluated. All three mutants (SpCas9-HF1, eSpCas9(1.1), and HypaCas9) demonstrated at least an 8-fold reduction in off-target cleavage at five of the six sites compared to WT SpCas9 (Chen et al., 2017). These experiments demonstrate enhanced specificity extending beyond the previously demonstrated PAM-proximal seed regions of the sgRNA sequence when employing the HypaCas9 nuclease. Reducing the interaction between non-target strand DNA and Cas9 could be a valuable strategy to improve the safety of this system in therapeutic applications.
In addition to engineering the Cas9 nuclease to improve specificity, guide sequences have also been modified. Truncated guide RNAs (trugRNAs) reduce off-target cleavage by as much as 5000-fold. By combining the trugRNAs with Cas9n, unintended genomic modification can be further reduced (Fu et al., 2014). The investigators hypothesized that by decreasing the sgRNA-DNA interface, off-target DSB induction would be reduced (Fu et al., 2014). As mismatches are tolerated at the 5’ end of the guide sequence (Hsu et al., 2013), the authors reasoned that the removal of 5’ nucleotides would not be detrimental to on-target activity (Fu et al., 2014). When testing a range of guide lengths (15, 17, 19, and 20 nt) in EGFP reporter assays, the group demonstrated that sgRNA lengths of 17–18 nt cleaved as efficiently as matched full-length control sgRNAs (Fu et al., 2014). When the investigators combined the trugRNAs with the Cas9n nickase approach, deep sequencing to determine mutation frequencies revealed that off-target mutation rates dropped below the detection limit while the on-target cleavage remained comparable to full-length sgRNA controls (Fu et al., 2014). Taken together, these experiments highlight the improved specificity of trugRNAs, especially when combined with the Cas9 nickase approach.
2.6.1. Engineering alternate PAM recognition sites
Though CRISPR-Cas9 is a versatile and tractable system for gene editing, targeting is limited by the availability of specific PAM sites (i.e., sequences required for Cas9 recognition of target DNA) (Leenay and Beisel, 2017). The widely used S. pyogenes Cas9 nuclease primarily requires the NGG triplet immediately distal to the guide sequence (Jinek et al., 2012), which occurs on average every eight base pairs in the human genome (Gasiunas and Siksnys, 2013). However, some genomic loci carry a paucity of GC nucleotides making targeting these areas difficult. Engineered Cas9 nucleases with altered PAM sequence requirements address this constraint (Anders et al., 2016; S. Hirano et al., 2016; Kleinstiver et al., 2015). For instance, mutation of the SpCas9 PAM-interacting domain (Anders et al., 2014; Gasiunas et al., 2012; F. Jiang et al., 2015; Jinek et al., 2014; 2012; Nishimasu et al., 2014) and subsequent bacterial selection against an NGA PAM target site revealed two variants demonstrating the greatest discrimination between the NGA and NGG PAMs: D1135V/R1335Q/T1337R and D1135E/R1335Q/T1337R (Kleinstiver et al., 2015). Further characterization of PAM site specificity for these two variants – referred to as VQR and EQR, respectively – revealed that the VQR variant preferred NGAG and NGCG PAMs while the EQR variant was specific for NGAG sequences only (Kleinstiver et al., 2015). When the researchers extended their studies to select against the NGC PAM sequence, they demonstrated that a variant carrying four substitutions (D1135V/G1218R/R1335E/T1337R – referred to as VRER) was highly-specific for NGCG PAMs (Kleinstiver et al., 2015). Testing their activities in human cells against endogenous genes demonstrated that the VQR variant modified sites carrying NGA PAMs with a frequency of 6–53% (Kleinstiver et al., 2015). The VRER variant displayed 5–36% modification at endogenous human sites with NGCG PAMs (Kleinstiver et al., 2015).
Recently, a unique approach to expanding genome coverage of CRISPR-Cas targeting in mammalian cells (termed “proxy-CRISPR”) was reported (Chen et al., 2017). The type II-B Cas9 derived from Francisella novicida (FnCas9) has higher specificity than SpCas9, however, it is not active at many genomic loci in human cells. It was proposed that certain chromatin contexts inhibited FnCas9 access, thus attenuating its activity at certain loci (Chen et al., 2017). To that end, the catalytically dead SpCas9 (SpdCas9), was targeted to sites proximal to the FnCas9 targets previously inhibited. The researchers demonstrated that SpdCas9 binding at one proximal site enabled increased FnCas9 cleavage considerably at the POR locus (10–11% indels versus 0% with FnCas9 alone). SpdCas9 binding at two proximal sites further improved FnCas9 function (28% indels). Similar increases in SpCas9 activity were observed when Chen et al. reversed the roles of the two CRISPR-Cas systems by employing proximal binding of catalytically dead FnCas9 (FndCas9). The reverse strategy revealed an increase from 0.7 to 11% indels induced by SpCas9 at the POR target. Further investigation indicated that the optimal distance between proximal dCas9 binding sites and the cleavage target ranges between 7 and 50 nucleotides (Chen et al., 2017). Because chromatin structure varies between cell types it will be important to evaluate the distance between binding sites in several cell types including pluripotent stem cells.
2.7. Off-target detection
As indicated above, for the CRISPR-Cas system to be useful for clinical applications, editing at unintended or off-target loci across the genome must be evaluated. Several different methods for evaluating post-CRISPR off target events have been devised and range from bioinformatic analysis to whole genome sequencing. Below are some of the most promising techniques used to date.
2.7.1. Cell-based methods to evaluate off-target cleavage
Genome-wide, unbiased identification of DSBs enabled by sequencing (GUIDE-seq) (Tsai et al., 2014) is a procedure that employs the capture of double-stranded oligodeoxynucleotides within sgRNA-guided nuclease cleavage to provide evaluation of genome-wide cleavage in situ. Following CRISPR-Cas treatment, tagged double-stranded oligos are delivered and inserted into DSBs. Tagged loci are identified via Next Generation Sequencing (NGS). Application of 13 guides in two human cell lines – U2OS and HEK293 – demonstrated a wide variety of off-target cleavage sites for each guide (ranging from zero to greater than 150 sites) (Tsai et al., 2016). In addition to the previously identified off-target sites for four of the guides, the data also revealed many previously unknown off-target loci distributed among exons, introns, and non-coding intergenic regions. Extending the analysis quantitatively identified the contributions of different variables (number, location, and type of mismatches to on-target sequence, PAM density, expression level, and genomic feature) to the variability of off-target cleavage sites. The investigators determined that off-target sites harbored as many as six mismatches (Tsai et al., 2016). Mismatches were better tolerated when positioned one-four base pairs distal to the PAM compared to those five-eight base pairs away (Tsai et al., 2016). Overall, the factors contributing the most to off-target site variability were the number, location within the guide sequence, and type of mismatch. Notably, the researchers observed positive linear correlations between the number of off-target mutations discovered and the indel mutation frequencies (Tsai et al., 2016). Direct comparison of data produced via GUIDE-seq to in silico off-target predictions generated by the MIT CRISPR Design Tool (Ran et al., 2013b) and the E-CRISPR software (Heigwer et al., 2014) indicated that the majority of sites recovered via GUIDE-seq were missed by the computational approaches (Tsai et al., 2016). Many of the missed sites harbored more than three or four mismatches, thus they were not considered by the E-CRISPR or MIT programs, respectively. To assist in data analysis using this method, a streamlined open-source software called the Python package (Tsai et al., 2016) and a Bioconductor package in R (Zhu et al., 2017) were developed and made available.
Another genome-wide in situ approach to map off-target cleavage at nucleotide resolution utilizes ligation of a biotin-labeled linker to DSBs followed by streptavidin enrichment and NGS analysis. This method is called breaks labeling, enrichment on streptavidin, and next-generation sequencing (BLESS) (Crosetto et al., 2013). Following nuclease-mediated DSB induction, biotin-labeled proximal linkers are ligated in situ, gDNA is extracted, fragmented, and captured on streptavidin beads. Free ends are ligated to a second linker and fragments released by digestion with I-SceI. Released fragments are amplified by PCR using linker-specific primers and sequenced. Genome-wide targeting specificity of SaCas9 following in vivo delivery of an AAV vector expressing Pcsk9-specific guides and SaCas9 into murine liver was assessed using this method (Ran et al., 2015). Targeted deep sequencing analysis of candidate off-target sites identified by BLESS did not demonstrate significant frequencies of indels in liver tissue (n = 3 animals) as assessed four weeks post-injection (2 × 1011 genome copies) (Ran et al., 2015).
An alternative to both GUIDE-seq and BLESS is the Breaks Labeling In Situ and Sequencing (BLISS) method (Yan et al., 2017). This approach enables direct labeling of CRISPR-Cas-induced DSBs in paraformaldehyde-fixed cells or tissues and on solid surfaces. BLISS can accommodate low-input samples via linear amplification of the labeled DSBs by in vitro transcription. DSB frequencies can be quantified through unique sample barcodes introduced in the amplification step. Next generation sequencing facilitates scalability and multiplexing. Comparison of BLISS to other genome-wide DSB assessment approaches (targeted next generation sequencing, BLESS, GUIDE-seq, and Digenome-seq) at two previously characterized loci (EMX1 and VEGFA) demonstrated that in addition to many of the same SpCas9-mediated off-target cleavage events recovered with the other methods, BLISS detected four and 27 new sites not found with the BLESS method evaluating EMX1 and VEGFA, respectively (Yan et al., 2017). Comparison with GUIDE-seq and Digenome-seq revealed fewer total novel off-targets (Yan et al., 2017). BLISS recovered fewer off-targets sites for the type V CpfI nucleases from Acidaminococcus sp. (AsCpfI) and Lachnospiraceae bacterium (LbCpfI) when compared to the number recovered from SpCas9 indicating CpfI cleaves with greater specificity than SpCas9 (Yan et al., 2017).
Many of the important studies evaluating CRISPR-Cas9-induced off-target cleavage are performed in immortalized cell lines. While this is an important initial step in determining safety and efficacy of this powerful technology, these cells are not particularly well-suited to disease modeling and therapeutic applications as prolonged culture can result in chromosomal aberrations and malignant phenotypes (Erez et al., 2003; Hurlin et al., 1991). Moreover, when evaluating safety in patient-derived cells, it is important to distinguish between variations due to the nuclease treatment and those occurring from the reprogramming process itself. To address this limitation, whole genome sequencing (WGS) was performed at 60X coverage in ten human pluripotent stem cell clones, six of which were generated with CRISPR-Cas9 genome editing (Veres et al., 2014). Three of the hPSC clones were treated with CRISPR-Cas9 targeting SORT1; an additional three clones were treated with CRISPR-Cas9 directed towards LINC00116. In each cohort, one clone carried the wild-type allele and two clones carried indels at the respective loci (Veres et al., 2014). The investigators identified a total of 24 off-target indels across the six CRISPR-treated clones as compared to the parental cell pool, only one of which occurred within the coding sequence of a gene (ZDHHC11) or expressed sequence of annotated non-coding RNA (Veres et al., 2014). None of the indels were within 100 nucleotides of predicted off-target sites based on sequence similarity (Veres et al., 2014), which suggests that the majority of variations between CRISPR-treated and parental clones are not caused by off-target cleavage, but rather reflect the reprogramming and expansion process (Gore et al., 2011; Hussein et al., 2011).
2.7.2. Cell-free off-target detection
In contrast to the previously discussed off-target detection methods, the Digenome-seq (digested genome sequencing) and circularization for in vitro reporting of cleavage effects by sequencing (CIRCLE-seq) approaches rely on WGS of cell-free genomic DNA digested in vitro using CRISPR-Cas9 (D. Kim et al., 2015; 2016; Tsai et al., 2017). Digenome-seq digests produce identical 5’ ends that align vertically at the cleavage site of a given guide RNA. Evaluation of two guides previously shown to induce off-target cleavage at high frequencies demonstrates Digenome-seq reproducibly detects off-target cleavage when frequencies are as low as 0.1% (D. Kim et al., 2015).
The most recently developed method for detection of off-target effects following CRISPR-Cas9 treatment is CIRCLE-seq (Tsai et al., 2017). Following in vitro digestion, adaptor ligation, and PCR amplification of sheard and recircularized DNA molecules containing Cas9 cleavage sites, off-target sites are sequenced with paired-end deep sequencing in a genome-wide manner. The nuclease-cleaved sequence reads were recovered with an estimated 180,000-fold enrichment rate relative to random background using gDNA from HBB-targeted K562 cells. Comparison to other cell-based off-target detection methods indicated many more off-target cleavage loci were recovered with CIRCLE-seq (Tsai et al., 2017). In some cases, one to three sites previously identified by the other methods were not recovered, however, upon repetition or sequencing with greater depth these discrepancies were accounted for (Tsai et al., 2017). A previous study employing WGS of CRISPR-Cas9-treated human iPSCs revealed a high-frequency off-target sites generated by a single nucleotide variation (L. Yang et al., 2014). Comparison of off-target events generated by six previously characterized sgRNAs in two cell lines, U2OS and HEK293 revealed a number of sites preferentially cleaved in one cell type over the other a subset of which carried non-reference sequence variants. These studies highlight the need for taking genetic background into account when assessing CRISPR-Cas9 modification in patient cells (Tsai et al., 2017; L. Yang et al., 2014).
The studies outlined in this section demonstrate the vast array of methods developed to address the important challenge of off-target cleavage detection in CRISPR-Cas treated cells and tissues. Several algorithms generated can predict potential sites, however, it is important to complement these assessments with unbiased evaluation of off-target cleavage in the intended cell types. In this regard, uncovering these events in patient-derived iPSCs prior to transplantation will be relatively straightforward, especially if employing whole genome sequencing for example. Regardless of the method selected, in vivo analysis of off-target cleavage still presents a challenge that warrants further study.
3. CRISPR delivery methods
Multiple approaches have been used to successfully deliver the CRISPR-Cas9 components to a cell. A thoughtful consideration is necessary to determine which approach is appropriate for the cell type of interest and the experimental design. Below we will discuss the different methods for CRISPR genome editing and the components that comprise the CRISPR-Cas system, as well as design consideration for HDR at a locus of interest.
3.1. In vitro delivery
3.1.1. Chemical transfections
Chemical transfections utilize lipid-based reagents or cationic polymers to effectively deliver DNA, RNA, and/or proteins to the nucleus of actively dividing cells. Although these reagents were very efficient for delivery of DNA to stably transformed cell lines historically, they suffered from poor efficiencies when treating stem cells, which limited their use (Tsvetanova et al., 2011). However, in our experience, recent products such as the Lipofectamine Stem Transfection Reagent have high transfection efficiencies (>50%) and low cellular toxicity (Enzmann et al., 1998). The advantage of chemical transfections is that no expensive instrumentation is needed and the protocols are straightforward.
3.1.2. Electroporation
Another common method for delivery of genome editing reagents to mammalian cells is electroporation. This technique utilizes a pulse(s) of high voltage to cause pore formation and membrane depolarization, which creates a gradient for DNA to enter the cell (Tsvetanova et al., 2011). Efficient transfections are possible, but electroporation of stem cells often results in a high level of cell death (Mohr et al., 2006). Also, specialized instrumentation, such as the NEON transfection system or Lonza’s Nucleofector, are required. While the Nucleofector has fixed settings and requires unique reagents for each cell type, the NEON transfection system allows for optimization using a single reagent for multiple cell types (Brees and Fransen, 2014).
3.2. In vivo retinal cell delivery
3.2.1. Viral delivery
Adeno-associated virus-mediated delivery systems have shown promise for delivery of CRISPR-Cas9 reagents (Ishizu et al., 2017; Ran et al., 2015; Refaey et al., 2017; Y. Yang et al., 2016). Recombinant AAV vectors persist mainly as episomes and are capable of transducing post-mitotic cells (Afione et al., 1996), and are already being used in clinical trials for gene replacement therapy in the retina (Bainbridge et al., 2008; Russell et al., 2017; Weleber et al., 2016). Multiple AAV serotypes present a broad viral tropism in vivo. For example, as shown in Figure 2 and described elsewhere (Auricchio et al., 2001; Lotery et al., 2003), capsid 5 shows an increased photoreceptor cell restricted tropism making it an ideal candidate for in vivo CRISPR based strategies targeting photoreceptor cells. For targeting inner retinal neurons and/or retinal ganglion cells in addition to photoreceptors, capsids 4 or 6 may be more appropriate (Figure 2). Yet, as indicated above AAVs have a relatively small packaging capacity (i.e., ~4.9 kb (Dong et al., 1996)), which limits the size and number of sequences that can be incorporated into a single virus (Ran et al., 2015). Moreover, DNA repair is highly regulated resulting in peak HR efficiencies between late S2 and early G2 phase making HR less efficient in post-mitotic cells (Su et al., 2015). Given the limited insert size and the predictably low HR correction efficiency, initial translational studies will likely be restricted to gene-knockout strategies (Burnight et al., 2017).
Figure 2. AAV serotype transduction comparison following subretinal injection in mouse retina.
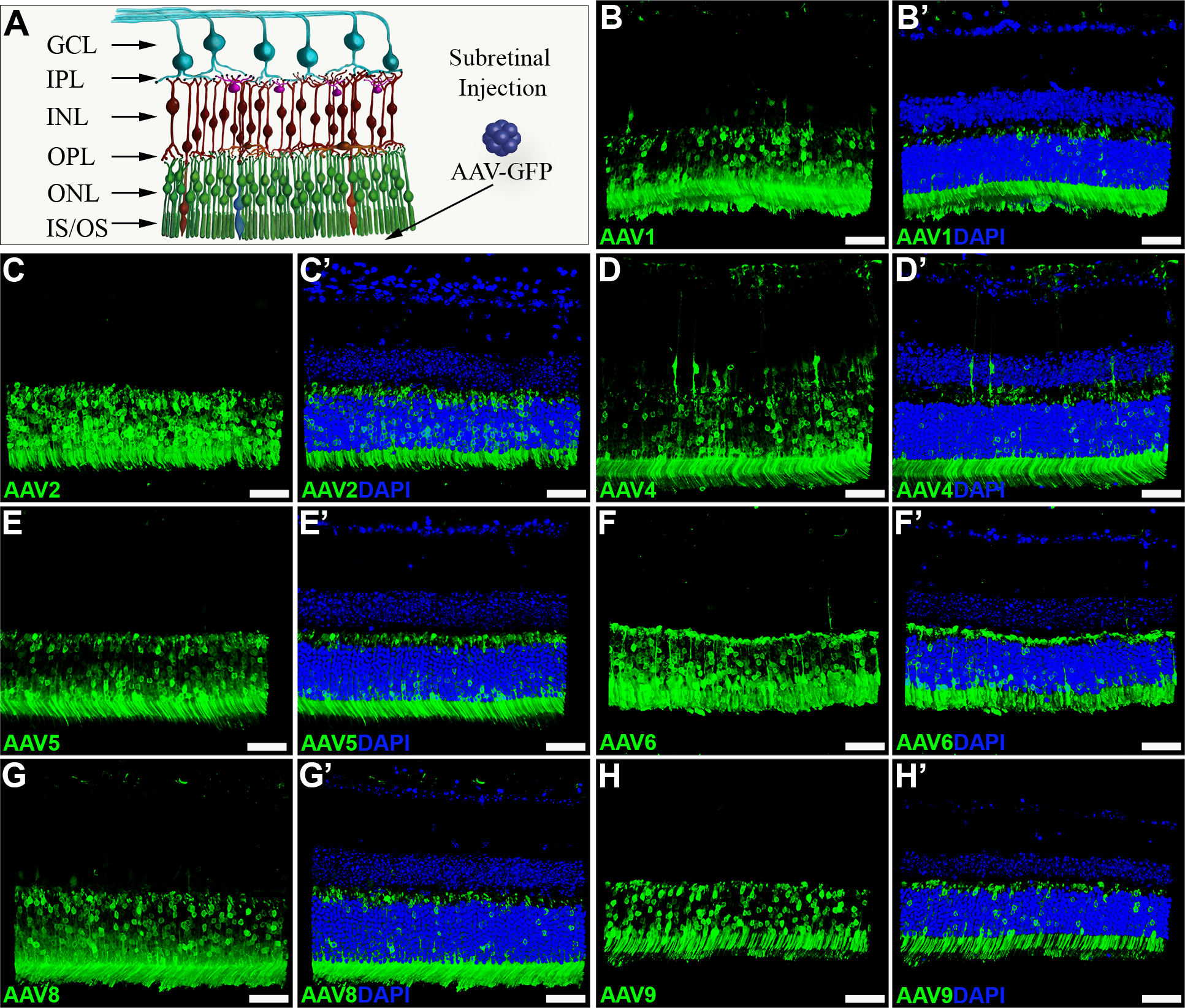
A) Schematic digram depicting the layers of the neural retinal and placement of AAV vectors in vivo. B-H) Immunohistochemical analysis of GFP expression (green) driven by seven different AAV serotypes two weeks post-subretinal injection. Representative z-stacks are shown for each of AAV1 (B-B’), AAV2 (C-C’), AAV4 (D-D’), AAV5 (E-E’), AAV6 (F-F’), AAV8 (G-G’) and AAV9 (H-H’). DAPI was used to visualize retinal nuclei. NFL – nerve fiber layer, GCL – ganglion cell layer, IPL – inner plexiform layer, INL – inner nuclear layer, OPL – outer plexiform layer, ONL – outer nuclear layer, IS/OS – photoreceptor cell inner and outer segments. Scale bars = 50 μm.
3.2.2. Nanomaterials for targeted delivery of CRISPR-Cas9
An alternative approach to viral vector mediated gene delivery is the use of engineered nanotechnologies for targeted gene delivery. Vectors of this type are composed of various materials including lipid, protein, and peptide-based nanoparticles and graphene nanotubes, as well as inorganic nanoparticles/polymers (Riley and Vermerris, 2017). These materials are often tunable and can be designed to overcome the challenge of viral vector-induced immunogenicity and payload limits (Iijima, 1991; Nayak and Herzog, 2010). For individuals diagnosed with inherited retinal disease, nanoparticle delivery systems that enable efficient retinal gene delivery (Adijanto and Naash, 2015), may prove to be a useful for in vivo CRISPR-Cas9 genome editing. These materials offer a safe vehicle, which prevents degradation of the CRISR-Cas9 system and a targeting ligand, which imparts tissue specificity (Zhen et al., 2017). For instance, recent studies have shown that AS1411, a G-quartet aptamer (i.e., a 29 base, single stranded DNA molecule), is capable of targeting protein cargoes into retinal cells in vivo (Leaderer et al., 2016) and as such may be useful for evaluating CRISPR mediated gene knockout.
4. Developing a CRISPR-Cas9 based strategy for retinal genome editing
For successful genome editing of retinal neurons, the Cas9 gene and a single guide RNA (sgRNA) must be delivered to the cell of interest using one of the approaches indicated above. These components can be delivered as either 1) plasmid DNA or 2) ribonucleoproteins (RNP).
1). Plasmid DNA
A typical CRISPR-Cas9 plasmid consists of two main expression cassettes: the RNA Pol III promoter (U6) driving expression of a chimeric, single small guide RNA (sgRNA) and a constitutive RNA Pol II promoter driving the expression of the Cas9 gene. Importantly, the 5’ 17–23 nucleotides within the sgRNA can be modified allowing for targeting of specific regions in the genome (Cong et al., 2013; Ran et al., 2013b).
2). Ribonucleoprotien (RNP)
The combination of protein and RNA constitutes a functional ribonucleoprotien or RNP. SgRNA transcripts can be generated via in vitro transcription or commercially ordered. Recombinant Cas9 protein is also commercially available. RNPs have two major advantages over plasmid based systems, 1) since no transcription or translation is necessary, the onset of action is immediate, and 2) their effects are transient compared to plasmid DNA, which can persist over multiple cell passages. As such for clinical applications this approach may be desirable.
4.1. Bioinformatic tools for sgRNA design
The next step in developing an effective CRISPR based genome editing strategy is to build sgRNAs designed to selective target the genomic locus of interest. Many free online programs have been developed to help researchers identify appropriate guide sequences. Some of these tools, such as CRISPOR (http://crispor.org) (Haeussler et al., 2016) and benchling.com allow for simultaneous consideration of multiple published algorithms, which are designed to assess off-target versus on-target predictions. Due to the drastic increase in predicted off-target sites, highly repetitive regions constitute poor sgRNA candidates and should be avoided.
4.2. HDR Strategies
As indicated above, in response to DSBs in DNA, genomic repair predominantly occurs through either NHEJ or HDR. For meaningful levels of HDR to occur, a donor template with homology to the targeted region must be simultaneously delivered with the CRISPR-Cas reagents. Both single-stranded oligodeoxynucleotide (ssODN) and double-stranded plasmid DNA have been used for this purpose. For SsODNs to be most effective they should be designed such that the intended genomic modification is located centrally and the DSB occurs within 10 base pairs of intended modification (L. Yang et al., 2013). Due to the fact that the length of gene conversion tracts in mammals is commonly less than 60 base pairs (Elliott et al., 1998), it has been reported that SsODNs should be designed with at least 40 base pairs of homology on either side of the target sequence (Ran et al., 2013b). Similarly, for plasmid-based constructs, it has been reported that the DSB should occur within 200 base pairs of the intended modification and homology arms of ~500 base pairs should be included (Byrne and Church, 2015). Due to the infancy of genome editing technology, most design considerations are based on experimental observation of a small sample sizes. As such, there is likely flexibility in the distance from DSB and intended genome modification as well as the required length of homology arms depending on the efficiency of correction at the locus of interest. In plasmid-based systems, drug resistance cassettes can be placed between the left and right homology arms allowing for subsequent post-modification selection (Enzmann et al., 1998). This approach is especially useful when developing genetically corrected clonal cell populations (i.e., excluding random plasmid integration, if the intended genomic modification is placed between the DSB and the drug resistance cassette, drug selection will result in the survival of only genetically corrected clones).
5. The CRISPR System In Vivo
As knowledge pertaining to the CRISPR-Cas9 technology continues to advance, our ability to accomplish genome editing in vivo in a wide variety of tissues for various applications will continue to improve. Genome editing in vivo provides the advantage of studying a physiological process or disease phenotype in the native tissue environment. Furthermore, in vivo work allows us to ask temporal questions relating to a gene or mutation, for example, we can study a given gene’s involvement at a specific point during development or the role of a specific genetic sequence in the development or amelioration of a disease. In vivo genome editing also allows for the investigation of cell-autonomous and non-cell-autonomous effects of specific disease-associated variants or changes in protein function. As discussed in detail below, few major challenges still exist before CRISPR-Cas9-based genome editing will be used to treat human disease in vivo, including preventing a patient’s immune response to CRISPR reagents, exogenous DNA or viral vectors, achieving tissue or cell specificity, preventing deleterious off-target modifications and reaching therapeutic levels of editing efficiency (i.e., especially if HDR in post-mitotic neurons is desired). Many groups have begun to overcome these challenges, and below we will outline the numerous in vivo studies that have been successfully conducted to date using animal models.
5.1. In vivo CRISPR-Cas9-mediated genome editing in the eye
As described above, CRISPR is proving to be a relatively facile and precise way to induce gene correction (Burnight et al., 2017; Wiley et al., 2015; Yiu et al., 2016). Rapid advances in the field, as described in the pre-clinical studies below, lend support to the notion that this method of gene editing could soon be used in clinical settings to treat inherited disorders of the eye (Tucker et al., 2014). The eye, unlike other tissues has several major advantages for testing of cutting edge treatments such as this. Anatomically the eye is a small, compartmentalized organ which is easily accessible and can be conveniently monitored using minimally invasive, well established techniques such as fundus examination and optical coherence tomography (Ochakovski et al., 2017). In addition, being a paired organ, one eye can be treated individually and the corresponding eye can be used as a contralateral control. As such, any adverse effects that may arise after treatment can be readily detected in the eye, treated locally and the trial can be immediately halted without further harm to the treated or future patients. For instance, in the unlikely event that a CRISPR induced off target modification were to occur in a cancer suppressor gene causing a tumor to be formed, the resulting tumor could be detected very early and destroyed via laser ablation prior to it inflicting significant harm. In areas outside the eye, like the brain for instance, tumors and other untoward events can be very difficult to detect until significant harm has been done.
Inherited eye diseases affect millions of people worldwide and have an immense socioeconomic impact (Sheffield and Stone, 2011). Collectively, retinal degenerative diseases are caused by the degeneration of retinal photoreceptor cells and treatments are currently limited. Ophthalmic genetics has improved tremendously in the last few years, in large part due to the advancements in molecular genetic testing technologies and precise clinical characterization of inherited retinal diseases (Sheffield and Stone, 2011; Stone et al., 2017). The ever-improving accuracy in identifying disease-causing mutations has the potential to lead to the development of more clinically relevant treatments, which may include CRISPR-Cas9 based genetic correction. Several approaches could employ CRISPR to treat retinal disease-causing mutations. For example, the mutation and disease causing allele itself could be targeted for deletion. Dominant monogenic diseases such as rhodopsin associated retinitis pigmentosa or Best disease would be likely candidates for this approach (Burnight et al., 2017). Alternatively, the mechanistic pathway known to be associated with disease could be targeted (e.g., inhibition of ER-stress via pathway targeting).
Several studies treating animal models of ocular disease using CRISPR-Cas9 in vivo are now emerging. For instance, a study using a rat model of severe retinal dystrophy caused by a single dominant point mutation in the rhodopsin gene (S334ter) (Bakondi et al., 2016) showed that the mutation could be corrected using delivery by a single subretinal injection of a plasmid expressing the sgRNA and electroporation to facilitate plasmid uptake. This animal model carries a mutated Rho gene which results in a truncated version of the rhodopsin protein, showing a similar phenotype to the human RHO protein misfolding mutations, leading to continual photoreceptor loss and blindness (McGill et al., 2012). By ablating the disease allele specifically, the phenotype could be corrected (Bakondi et al., 2016). In a similar series of experiments, we were able to demonstrate that following subretinal injection of a single AAV vector carrying SaCas9 and a Pro23His mutant rhodopsin specific sgRNA, we could selectively induce in del formation in the disease causing allele and in turn prevent translation of the mutant disease causing gene (Burnight et al., 2017).
Recently, CRISPR-Cas9 was employed to target the most frequent mutation contributing to CEP290-associated Leber congenital amaurosis (LCA), a deep intronic variant in intron 26 of CEP290 (c.2991+1655 A>G) (Ruan et al., 2017). The intronic mutation (referred to as IVS26) creates a splice site that results in inclusion of a cryptic exon and in-frame termination codon (Hollander et al., 2006; Perrault et al., 2007). To explore the feasibility of in vivo targeted intronic (and thus IVS26) sequence removal, the investigators generated a dual AAV system expressing a pair of sgRNAs and the SpCas9 in separate vectors. As proof-of-principle, the investigators packaged sgRNA pairs and SpCas9 into AAV5 and co-injected each vector subretinally into 8–10 week old C57CL/6J mice. Next generation sequencing analysis of treated retinas (n=4) four weeks post-injection revealed 7.5% - 26.4% of sequences carried genomic deletion of intron 25 sequences. These experiments supported the feasibility of targeted genomic deletion in murine photoreceptor cells using CRISPR-Cas9 (Ruan et al., 2017).
An alternative to correction of disease-causing mutations, is employing a CRISPR-Cas-mediated gene-independent approach. Specifically, as in RP, the loss of cone photoreceptor cells is secondary to loss of rod photoreceptor cells, targeting non-disease specifc genes in an attempt to preserve rods, even if non-function maybe beneficial. To that end, AAV-based CRISPR-Cas9 was recently employed to disrupt the gene Nrl, which is required for maintanence of rod cell fate, in postmitotic murine photoreceptors (W. Yu et al., 2017). SgRNA- and SpCas9-expressing AAV8 vectors were delivered subretinally to three mouse models of retinal degeration each exhibiting an initial phase of rod degeneration followed by secondary cone loss (Chang et al., 2007; Lem et al., 1999; T. Li et al., 1996). AAV-Nrl-CRISPR vectors were administered both prior to and after initiation of rod photoreceptor cell death. Importantly, retinae in mice treated at both times with CRISPR-Nrl vectors were significantly thicker and demonstrated better cone ERG responses than those of control mice, albeit to a lesser extent in mice treated at the later timepoint. These results persisted for up to four months of age. Although the translatability of NRL is questionalable, that is in humans mutations in this gene cause diseases ranging from recessive enhanced S cone syndrome (Littink et al., 2018) to autosomal dominant retinitis pigmentosa (Gao et al., 2016), the findings do highlight the potential therapeutic benefit of delaying rod photoreceptor cell death and in turn preserving cone photoreceptor cell health and function by using a CRISPR-Cas9 based disease gene independent approach (W. Yu et al., 2017).
Other approaches for disease alleviation that could be employed to target known disease mechanisms include a recent study in which the authors treated one of the main mechanisms associated with age-related macular degeneration (AMD) (K. Kim et al., 2017). As we discuss in further detail below, AMD is a multifactorial and complex disease with several known genetic risk factors as well as many contributing environmental variables. Wet AMD is associated with an increased vascularization caused in part by angiogenic cytokines such as VEGFa (Homayouni, 2009; Leung et al., 1989). The most effective treatment currently available for wet AMD is intravitreal administration of anti-VEGF agents that work to inhibit neovascularization, remove fluid from the retina, and slow disease progression (Schmidt-Erfurth et al., 2014; Stone, 2006). CRISPR-Cas9 can be used to locally reduce the Vegfa protein expression levels in vivo (K. Kim et al., 2017). When purified Cas9-ribonuclease proteins and sgRNA targeting the Vegfa gene were delivered subretinally, in combination with a transfection reagent, into a mouse model of AMD (laser-induced choroidal neovascularization (CNV)) targeted inactivation of Vegfa was specifically detected in the RPE, which resulted in a 58% reduction in CNV formation (Schmidt-Erfurth et al., 2014).
Each of the studies outlined here has advanced its respective field by describing novel gene editing systems to study and treat complex diseases in vivo. While these studies collectively show the excellent progress of the field in the use of CRISPR-Cas9 technology for treatment of retinal disease in vivo, there is still a significant amount of work to be done before this technology directly used for clinical applications. In the next section we will discuss the challenges toward therapeutic use of CRISPR in vivo.
5.2. Challenges toward therapeutic use of CRISPR-Cas9 in vivo
As indicated above, for patients with inherited retinal degenerative disorders such as retinitis pigmentosa who present early in the course of their disease, the CRISPR-Cas9 technology could, in theory, provide a viable treatment option for preventing cell dysfunction and death. This technology would be especially useful for genes that are too large to be packaged into clinically useful vector systems or for disease-causing variants in genes for which overexpression from heterologous promoters is known to cause cytotoxicity (Burnight et al., 2014; Seo et al., 2013). In the latter situation, CRISPR-Cas9 based correction would allow the gene to remain under control of the endogenous regulatory elements. Another attractive feature is that the coding sequences of the CRISPR-Cas system are small enough to be packaged into clinically-approved AAV gene therapy vectors (Friedland et al., 2015; Hung et al., 2016; E. Kim et al., 2017; Ran et al., 2015) for in vivo delivery. However, several challenges related to both off-target modification and on target efficiency following in vivo delivery remain to be solved before moving this technology into the clinic.
As already mentioned, delivery to the intended cell types can be achieved by using one of the various AAV serotypes available (Figure 2). For photoreceptor cell specific diseases such as retinitis pigmentosa, the levels of gene function and number of cells that need to be genetically corrected in order to slow disease progression and retain useful vision have not been well-defined and will likely need to be determined empirically for each gene. Likewise, the point in disease course in which the treatment can be administered and have a chance to be effective is also currently unknown. Regardless, further studies focused on evaluating efficacy and safety of CRISPR-based in vivo treatments are needed. This is critically important considering that photoreceptor cells are post-mitotic, which means that AAV vectors can persist for months to years after delivery thereby increasing the potential for off-target cleavage.
A recently published letter to the editor in Nature Methods raised concern that CRISPR-Cas9 can induce high levels of off-target cleavage in the rd1 mouse model of retinitis pigmentosa (Schaefer et al., 2017). In this study, whole genome sequencing demonstrated that two CRISPR-Cas9-treated mice had high numbers of single-nucleotide variants (SNV) and indels compared to a single control mouse from the same inbred strain, FVB/N. Several groups (S.-T. Kim et al., 2017; Lareau et al., 2017; Wilson et al., 2017) quickly criticized this study because the on-target site and the sites of off target mutations had very little sequence homology and most of the latter lacked a PAM sequence – both of which are at odds with the extensive published literature on CRISPR-Cas9’s mechanism of action (Jinek et al., 2013b; Mali et al., 2013b; Cong et al., 2013; Sander and Joung, 2014). Concerns were also raised about the small number of mice used in the study (one control and two CRISPR-Cas9-treated mice) and the lack of appropriate controls (Jinek et al., 2013a; Mali et al., 2013a; Sander and Joung, 2014). Studies of human family trios (both parents and one offspring) show that offspring have dozens of new germline mutations that are not present in either parent, and closely related individuals will therefore harbor many more shared novel mutations (and shared departures from the consensus sequence) than they share with less closely related individuals (Conrad et al., 2011). The concern raised about this study was that the pattern of variants observed between the 3 mice was most consistent with the possibility that the two CRISPR-Cas9-treated mice were in fact genetically more closely related to each other than the control mouse (S.-T. Kim et al., 2017; Lareau et al., 2017). In other words, it is possible that the mice in the two groups were genetically different before CRISPR-Cas9 treatment; and it would therefore seem that more studies are warranted to investigate the concerns raised in by the authors. (S.-T. Kim et al., 2017; Lareau et al., 2017; Wilson et al., 2017). This controversy also highlights the need for rigorous evaluation of off-target cleavage events in pre-clinical models before moving forward with therapeutic applications in vivo. One alternative to animal models for this purpose would be to pre-screen CRISPR-Cas reagents for the targeted loci using hIPSC derived retinal cells from the patient for whom the treatment is intended. This would allow researchers to test the efficacy and safety of CRISPR based treatments prior to being administered in vivo.
In addition to off-target events, a major challenge associated with the effective use of CRISPR based genome editing for treatment of retinal degenerative blindness in vivo is the relatively low efficiency of on-target editing. Although the in vivo studies reported to date have been encouraging, the efficiency of CRISPR induced on-target modifications in photoreceptor cells has been quite low. For instance, subretinal injection of an AAV vector carrying SaCas9 and a sgRNA targeting mutant rhodopsin induced indel formation at an efficiency of approximately 3.9%, which in this case would be well below the therapeutic threshold required to prevent disease progression (Burnight et al., 2017). Two important considerations pertaining to on-target efficiency are: 1) the efficiency of the clinical delivery method used; and, 2) the efficiency of the CRISPR reagents themselves after delivery. Although AAVs are an excellent vehicle for transfer of genomic information to the retina, the efficiency of transduction is well below 100%. For instance, as shown in Figure 3 at 1-week post-infection, less than 50% of the photoreceptor cells targeted with AAV5-GFP (one of the most efficient vectors for photoreceptor selective infection in human) were transduced (Wiley et al., 2017). In our hands the average NHEJ efficiency obtained in HEK293T cells in vitro ranges from ~4% to ~38% (Figure 3). Thus, with current technology one should expect no more than 0.2X correction using an AAV-delivered CRISPR-Cas9 therapeutic vector to the human retina. This calculation suggests that new strategies will be needed to increase the number of transduced photoreceptor cells and on-target correction efficiency, without increasing off-target events, before this approach will be helpful for most inherited retinal diseases.
Figure 3. Efficency of CRISPR based genome editing.
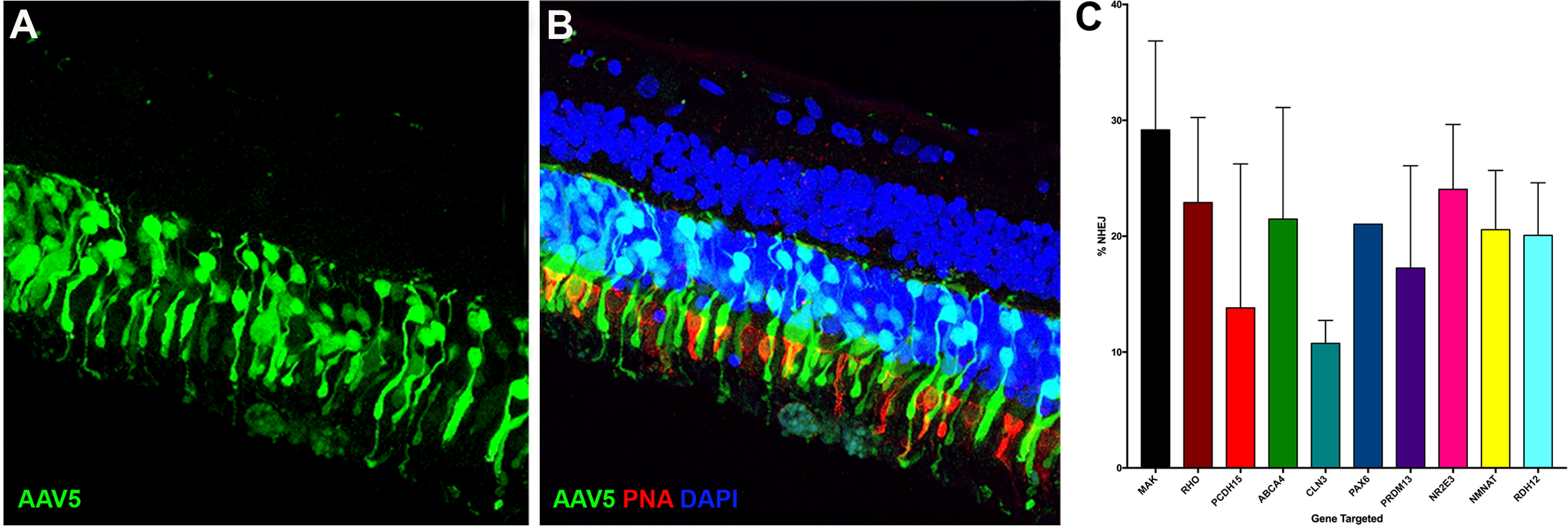
A-B: Immunohistochemical analysis of GFP expression (green) following transduction of human retinal explants with AAV5-GFP at 1 week post-subretinal delivery. Unlike in vitro HEK293 transfection efficiency, which is near 100%, AAV5 based gene delivery vectors typically transduce less that 50% of the photoreceptor cells targeted. C: In vitro NHEJ efficiency. Of 40 sgRNAs targeting ten independent genes associated with inherited retinal degenerative disease, an average NHEJ efficiency of 20.6 ± 1.3% NHEJ was detected.
6. In vitro applications of CRISPR therapeutics
Although the tractability of the CRISPR-Cas9 system presents many exciting opportunities for developing genetic therapies for inherited retinal disease, such as retinitis pigmentosa (Burnight et al., 2017) this system is especially well-suited for in vitro applications. In particular, the emergence of the CRISPR-Cas9 system has made it possible to efficiently correct a patient’s own skin derived iPSCs prior to differentiation, where corrected clones can be rigorously screened to ensure that a transient treatment has restored gene function without inducing adverse off-target events. This has opened the door for disease modeling of rare disorders, regardless of patient ethnicity (i.e. genetically corrected cells from the patient can be used as isogenic controls), and autologous cell replacement.
6.1. CRISPR-Cas9-mediated gene editing in Mendelian retinal disease
We recently employed CRISPR-Cas9 genome editing to develop strategies for targeting and correction of three major classes of inherited retinal degeneration-causing mutations: 1) exonic, 2) deep intronic splice site and 3) dominant gain-of-function mutations (Burnight et al., 2017).
To demonstrate utility of the CRISPR system for correction of exonic mutations, we targeted an exonic Alu-insertion mutation in the MAK gene (the leading cause of retinitis pigmentosa in individuals of Jewish ancestry) (Tucker et al., 2011b). Briefly, guides were designed using the CRISPR Design Tool (crispr.mit.edu) and genomic cutting efficiency was evaluated in HEK293T cells before employing the most efficient guide (which produced 31% indels) in patient-derived iPSCs. We co-delivered a plasmid expressing the guide and the human codon-optimized SpCas9 and a plasmid carrying donor repair sequence and a puromycin-selection cassette flanked by ~500 bp homology to the MAK target locus. Following selection and clonal expansion, we recovered six iPSC clones in which one allele was corrected. Expression of MAK transcript and protein was restored in one of the six clones.
To demonstrate utility of the CRISPR system for correction of intronic non-coding variants, we targeted the most common mutation contributing to CEP290-associated Leber congenital amaurosis (LCA), a deep intronic variant that creates a cryptic splice site in intron 26 (IVS26) (Hollander et al., 2006). To remove the mutation, we employed single- and dual-guide strategies in patient specific iPSCs homozygous for the IVS26 disease-causing mutation (Burnight et al., 2017). We co-delivered plasmids expressing guides and a human codon optimized SpCas9 and T2A-GFP tag to facilitate clonal selection. The mutation was removed in greater than 50% of clones generated with the dual-guide strategy. Importantly, both transcript and protein expression were restored in cells treated with the IVS26-targeted CRISPR-Cas9 reagents. We extended these studies by employing the smaller S. aureus Cas9 to achieve correction via both nucleotide removal via NHEJ and nucleotide correction via homology-dependent repair. Upon delivery of a plasmid expressing two guides and the SaCas9 we observed an increase in wild-type CEP290 transcript. To achieve gene correction via homology-directed repair, we co-delivered a plasmid expressing two guides and the SaCas9 and a donor plasmid carrying wild-type sequence and a puromycin selection cassette. Following selection and clonal expansion, five of twelve puromycin resistant colonies carried correction on at least one allele. RT-PCR analysis revealed that one clone underwent biallelic correction. Importantly, protein expression was restored in five clones following CRISPR-Cas9 treatment. Collectively, we demonstrated that both single- and dual-guide strategies, delivered with either SpCas9 or SaCas9, could be used to remove a deep intronic cryptic splice site mutation and restore expression in CEP290-associated LCA patient-derived iPSCs (Burnight et al., 2017).
Finally, to demonstrate utility of the CRISPR system for correction of dominant gain of function mutations, we developed an allele-specific CRISPR-Cas9-based genome editing strategy, targeting the dominant gain-of-function Pro23His mutation in the rhodopsin gene (the most common cause of dominant retinitis pigmentosa (Stone et al., 2017)). When we surveyed the locus for available PAM sequences, we discovered that the C to A transversion laid within the seed region of a S. aureus target which we employed to modify the mutant His allele specifically. When compared to a matched control guide targeting the wild type Pro allele, we observed a 22-fold decrease in indel formation in cells that only carried the Pro allele. In addition, we also evaluated homology-directed repair for correction of the Pro23His mutation in patient-derived cells. We delivered a donor plasmid carrying wild-type sequence and a puromycin selection cassette along with a plasmid expressing a non-allele-specific guide and SpCas9. Following selection and clonal expansion, we observed genomic correction in five of eleven resistant clones.
In a similar series of experiments, iPSCs generated from a deaf patient with compound heterozygous mutations in the gene MYO7A (leading cause of type 1 usher syndrome, which causes early onset deafness and later onset retinitis pigmentosa), CRISPR-Cas9 correction of one allele was achieved through electroporation with a plasmid expressing the sgRNA and human codon-optimized SpCas9 and a 150-bp ssODN. After clonal selection and screening, the investigators recovered three of 45 corrected clones (~7%). As this group was interested in hearing loss, hair-like cells were generated from both CRISPR corrected iPSCs as well as from the asymptomatic parent and an unaffected donor. Corrected iPSCs were shown to have restored organization of stereocilia-like protrusions. Likewise, the electrophysiological function of the CRISPR corrected cells was restored to levels similar to that of control cells (asymptomatic father of patient and unaffected control donor). (Tang et al., 2016).
Taken together, these studies illustrate the broad range of mutations that can be genomically corrected using both S. pyogenes and S. aureus Cas9 orthologs in patient specific iPSCs (Burnight et al., 2017; Tang et al., 2016). It not only demonstrates the utility of the NHEJ approach but it also demonstrates the power of the HDR based gene correction strategy (using both plasmid and SSODNs) which will be difficult to achieve in post-mitotic neurons such as photoreceptor cells in vitro.
6.2. CRISPR-Cas9-mediated gene editing and genetically complex disease
Complex diseases are those that are caused by many contributing factors, often resulting from the combined effects of multiple genetic factors, lifestyle, and/or environmental factors (eg. heart disease, cancer, and diabetes). The degree of genetic involvement in complex diseases is inherently more challenging to identify, therefore the development of effective disease treatments is more difficult to accomplish, which often results in a lack of treatment options.
Age-related macular degeneration (AMD) is an excellent example of complex disease in the eye. AMD is a retinal degenerative disorder in which the ultimate loss of photoreceptor cells within the macula results in loss of central vision. Loss of choroidal endothelial cells (CECs) and retinal pigment epithelium (RPE) precedes photoreceptor cell loss, making CECs and RPE cells the focus of much AMD research (Chirco et al., 2017; Songstad et al., 2015; 2017). As one of the most common causes of irreversible vision impairment in developed countries, AMD affects approximately 11 million people in the United States alone (Pennington and Deangelis, 2016; Wong et al., 2014). As the population ages, the prevalence of AMD continues to steadily rise; therefore, identifying and understanding the genetic contributions to AMD will aid in our understanding of disease pathophysiology and promote the development of treatment options for the millions of individuals affected by this disease.
To date, 34 genetic loci have been associated with AMD risk (Fritsche et al., 2016). Among these, the strongest genetic associations include: 1) a single nucleotide polymorphism (SNP, rs1061170) in the CFH gene, which results in the replacement of a tyrosine with a histidine at amino acid 402 (Y402H) of the complement factor H (CFH) protein, and 2) a locus on chromosome 10q, which contains a coding region polymorphism and an insertion-deletion in the 3’ untranslated region of the gene ARMS2, and a promoter polymorphism and two synonymous variants in the first exon of the gene HTRA1. Despite the advances that have been made in understand the role of CFH and the 10q locus in AMD pathophysiology, surprisingly little is known about how variants in these loci contribute to disease risk.
Modifying or correcting alleles associated with AMD risk, since these are risk loci and are not direct disease-causing variants, may not be worthwhile at this time. However, the ability to assess genotype/phenotype relationships in human cells by studying the effects of specific mutations or polymorphism in vitro may provide new insight into disease mechanisms and uncover important disease-causing variants. With the advent of CRISPR-Cas9 technologies, we can now create specific disease-associated variants to study their contribution to disease phenotypes in a cell-specific manner. For example, the genome of CEC lines, primary cells, or iPSC-derived CECs without a given disease-associated variant can be edited using CRISPR-Cas9 to subsequently carry the variant(s) of interest. Alternatively, the reverse scenario can be carried out, in which a cell line harboring a risk allele from a patient with AMD is corrected using CRISPR-Cas9 and the effect on cellular phenotype is evaluated. This type of editing becomes particularly useful when studying a complex disease, because as indicated above, it allows one to generate isogenic control lines, which are essentially identical to the experimental cell lines with the exception of the mutation of interest. Since the genetic backgrounds of the cases and controls are the same when using this approach, and the variant of interest is the only difference between cell lines, any observed phenotypic differences are more likely to be a result of the altered gene or locus (presuming of course that no deleterious off target cleavage events were created).
As an example, one could use CRISPR-Cas9 to introduce the Y402H variant in the genome of a CEC line or iPSC-derived CECs to further understand the role of CFH in AMD pathophysiology. Since the primary function of CFH is to protect the cells against complement-mediated injury, the cells’ response to complement exposure can be compared to that of their isogenic controls. Furthermore, since ARMS2/HTRA1 are in linkage disequilibrium, CRISPR-Cas9 provides a promising tool to study mutations in these genes individually, in order to tease out which of these genes or variants may be important in AMD pathophysiology. For example, CRISPR-Cas9 could be used to generate four separate cell lines, each harboring one of the four variants in the ARMS2/HTRA1 risk locus. Combinations of variants can also be achieved in a given cell line using CRISPR-Cas9 to determine if more than one variant is required to cause a disease phenotype. In addition, genetic confounders, such as the highly prevalent high-risk CFH allele, which have added to the challenge of studying the ARMS2/HTRA1 locus in the past, can also be more easily avoided using the CRISPR-Cas9 method of gene editing.
7. CRISPR corrected iPSCs for autologous cell replacement
7.1. Opportunity and recent advances
In severe or advanced cases of retinal degeneration, in which patients have often lost the majority of their photoreceptor cells by the time they receive a diagnosis, gene augmentation or gene editing based treatment approaches are not likely to be effective (i.e., there are no cells left to treat). Photoreceptor cell replacement will ultimately be required if these patients are to regain visual ability.
The use of stem cells and their progeny has long been established as a viable approach for treatment of retinal degenerative blindness. Over the past decade numerous studies focused on the use of stem cell derived RPE cells for the treatment of AMD have been published (Koss et al., 2016; Mandai et al., 2017; Nommiste et al., 2017; Plaza Reyes et al., 2016; J. Wang et al., 2016b).
Importantly, the promising results afforded by these animal studies has recently budded into the first group of clinical trials focused on treatment of patients with advanced AMD. The first of these studies suggested that subretinal injection of embryonic stem cell-derived RPE in patients with AMD was relatively safe (Schwartz et al., 2015; Song et al., 2015).
Meanwhile, the latest advances in retinal cell differentiation protocols have enhanced our collective ability to produce sufficient numbers of stem cell-derived photoreceptor cells for retinal transplantation (Chao et al., 2017; Kundu et al., 2017; McGill et al., 2017; Tucker et al., 2013a; Wiley et al., 2016; Worthington et al., 2017). Transplantation of stem cell derived retinal progenitor cells have been shown to form functional synapses with host bipolar cells (Mandai et al., 2017; Singh et al., 2013; Tucker et al., 2011a) a marker of functional progress that had previously been elusive. Importantly, cell sorting to select non-proliferative cells and/or cells expressing desired retinal markers has recently proven to be beneficial in retinal transplantation (Lakowski et al., 2015; Shao et al., 2017). This process could be especially important given that retinal cells have now been shown to express differential immunogenetic markers compared to other cell types (Zhao et al., 2015).
7.2. Autologous cell replacement
In the past five years, two independent teams have directly compared the safety and efficacy of human embryonic stem cell (ESC)- and iPSC-derived retinal cell transplantations in animal models of retinal degeneration (Barnea-Cramer et al., 2016; Riera et al., 2016). In each study, cell integration and functional improvement occurred regardless of cell source, while the degree of photoreceptor-like cell integration depended on the number of surviving cells. However, the xenogeneic nature of these studies necessitated animal immunosuppression, removing the host immune response as a consideration. Although in the normal eye the blood-retinal barrier (BRB) does afford the retina some degree of immune privilege, when the retina is diseased circulating immunomodulatory factors have been shown to have easy access (Chinnery et al., 2012; Mullins et al., 2007; 2012; Rutar et al., 2010). In fact, the subretinal space has been shown to become pro-inflammatory in the face of photoreceptor and RPE degeneration (Ambati et al., 2013; Anand et al., 2003; Chinnery et al., 2012; Mullins et al., 2012; Rutar et al., 2010; Tarallo et al., 2012; Whitcup et al., 2013) and it is well established that inflammation plays an important role in the pathogenesis of both RP and AMD (Ambati et al., 2013; Anand et al., 2003; Tarallo et al., 2012; Tucker et al., 2013a; 2011b; 2011a; 2013b; Whitcup et al., 2013; Yao et al., 2011). Major histocompatibility complex (MHC) matching could help to mitigate concerns of immune rejection (Sugita et al., 2016), however years of MHC-matched organ and tissue transplantation in human demonstrates that this process is imperfect, and that the degree of MHC matching dictates the amount of systemic immunosuppression needed. As immune suppression leads to dramatically increased risk of infection and a significant reduction in quality of life, transplantation of cells that are immunologically entirely recognized as “self” would be a major advance. Taken together, these results suggest that autologous iPSC-derived retinal cells are likely to be the ideal cell type for the treatment of retinal degeneration. Although patients with complex disease such as AMD may benefit from use of native patient specific iPSCs (i.e. used without genetic manipulation), for individuals with Mendelian disorders caused by genetic mutations in known disease causing genes, gene correction should be performed before cellular differentiation and autologous cell transplantation. As discussed above, there have been several recent studies showing that such correction can be accomplished via CRISPR-based genome editing (Burnight et al., 2017; Hung et al., 2016; Ruan et al., 2017; Yiu et al., 2016), which, unlike traditional methods of gene augmentation, allow the host gene to remain under control of its endogenous regulatory elements.
7.2.1. Selecting Corrected iPSCs
In most situations, for autologous cell replacement a homogenous population of CRISPR-corrected iPSCs will be necessary. If a particular experimental approach results in a high percentage of the desired genomic modification, distributing the treated cells over multiple wells of a tissue culture plate and manually selecting and expanding iPSC clones may be sufficient to identify a homogenous subgroup. Yet, for patient-specific iPSCs homologous recombination typically occurs in less than 1% of the population of cells treated, which makes an iterative enrichment approach time and cost prohibitive (Byrne et al., 2015; Miyaoka et al., 2016).
To overcome this barrier, researchers have developed techniques to clonally select cells using FACS or reporter genes. For FACS, successful protocols require specific media, small molecules, and feeder layers to support single cell iPSC survival (Burnight et al., 2017; Byrne et al., 2014). After sorting, only a fraction of cells survive and the remaining wells must be screened for the intended genomic modification increasing the experimental time. Alternatively, a reporter cassette can be included in the homology directed repair construct allowing for the subsequent selection of corrected clones (Burnight et al., 2017; Byrne et al., 2015; Hou et al., 2013; Merkle et al., 2015). The reporter gene can encode for a fluorescent protein, allowing later separation of corrected clones, or for resistance to a specific antibiotic, which would then be added to the cell culture media (Giacalone et al., 2018). However, fluorescent proteins can be cytotoxic and fail to persist long-term (Ansari et al., 2016), so most clinical applications are better served by drug selection, which allows for positive, clonal selection, and subsequent expansion of corrected iPSCs. Ultimately, the use of selection reduces the difficulty of obtaining genome-edited cells because the desired modification is coupled to the selection cassette.
One thing to consider when inserting foreign sequences into the genome of an iPSC is that the targeted incorporation of selection cassettes has been shown to have short range and long-range effects on transcription (Meier et al., 2010; Pham et al., 1996; Zou et al., 2011). If placed near proto-oncogenes, for example, selection cassette integration may lead to insertional mutagenesis (Hacein-Bey-Abina et al., 2003). Other undesirable outcomes of inappropriate placement include the silencing of transgenes or simply ineffective selection (Rivière et al., 2012). Therefore, the ideal strategy is one designed to remove the reporter following use via methods such as the CRE/Lox system (Burnight et al., 2018; Giacalone et al., 2018).
An alternative approach for selection allows researchers to simultaneously genetically modify a sequence of interest and insert a reporter gene at an alternative location. This can be accomplished by delivering the necessary components to modify a sequence of interest while also delivering the necessary components to insert a reporter gene at a fixed locus (Mitzelfelt et al., 2017). In certain situations, this method has been shown to be more efficient than other selection techniques, with as much as 40% of correction being “precision” events (i.e. HDR vs. NHEJ) (Mitzelfelt et al., 2017). To define a locus suitable for the reporter cassette, experts in the field have been working together to identify genomic regions that when disrupted, are unlikely to cause discernable phenotypic effects. Also known as “safe harbors,” three of these regions that have been tested most extensively are AAVS1, CCR5 and human ROSA26 (Sadelain et al., 2011). AAVS1 in particular has been most rigorously evaluated (Cerbini et al., 2015; Coluccio et al., 2013; Holkers et al., 2014; Hong et al., 2017; Maggio et al., 2014; J. Wang et al., 2016a), emerging as the gold standard of “safe harbors.” However, AAVS1 does not meet the most recent criteria recommended for a truly safe genomic selection region. In order to do so, a locus must be at least 50 kilobases from the 5’ end of any gene, 300 kilobases from cancer-related genes, 300 kilobases from microRNA, outside of the gene transcription unit and outside of ultra-conserved regions (Sadelain et al., 2011). As we learn more about identifying and evaluating “safe harbors loci,” additional “safe harbor” options are likely to emerge. For example, we still know very little about whether or not “safe harbors” are differentially expressed by diverse cell phenotypes or whether they participate in critical DNA folding events. Recent technological advancement has shown that topologically associating domains, which are used to understand nuclear organization, can vary by cell-type, which has implications for changes in long-range interactions (Cubeñas-Potts and Corces, 2015). Furthermore, new genes, particularly those related to cancer, are discovered frequently, which narrows the pool of appropriate loci. Over time, fully validating the safety of using any of these genomic regions for selection could be costly and laborious, and ultimately, the data may necessitate the excision of the reporter gene.
7.2.2. Biomaterials and retinal tissue engineering
Despite the cell therapy advances described above, survival and integration of stem cell-derived retinal cells remains a challenge (Santos-Ferreira et al., 2016). There is strong evidence to suggest that visual improvement is correlated to the number of engrafted cells (Barnea-Cramer et al., 2016; Tucker et al., 2011a), highlighting the need for a delivery vehicle that protects CRISPR-corrected, autologous cells during and after transplantation. It is widely accepted that these challenges can be addressed using polymeric biomaterials, and the past few years of research in the area have increased our collective understanding of the optimum properties of such a material. For example, new polymer chemistries have been introduced, like the use of parylene-C and polyimide films for the support of RPE cells, which have been shown to be safe and effective in the porcine subretinal space (Brant Fernandes et al., 2016). On the other hand, transplantation of RPE-seeded polyimide films in the rabbit subretinal space caused de-pigmentation over time despite very little immune response to the polymer alone (Ilmarinen et al., 2015). In addition to these new polymers, variations on traditional degradable films have also been introduced, including using vitronectin to coat poly(caprolactone) (PCL), which increased RPC attachment and helped drive differentiation (Lawley et al., 2015). In addition, PCL and poly(lactic acid) (PLA) copolymers have been verified as an effective RPE substrate (Sorkio et al., 2015). Importantly, this study indicates that tuning the degradation profile of such materials by adjusting the ratio of each polymer is feasible; an important aspect of design given the acidic byproducts produced as polyesters degrade.
All of the above-mentioned synthetic degradable polymers, which are the most common materials considered for retinal cell delivery to date, are relatively stiff compared to the retina. Fortunately, the recent characterization of retinal modulus (10–20 kPa) has been accompanied by efforts to tune biomaterials to be mechanically matched to the subretinal space (Worthington et al., 2014). Interestingly, differences in modulus near the retinal range do not appear to influence the survival of photoreceptor cells encapsulated in hyaluronic acid (HA)-based hydrogels. Rather, HA itself is thought to interact with CD44 receptors and activate mTor pathways to promote survival and integration in the mouse subretinal space (Ballios et al., 2015). The same materials, when modified with pro-survival proteins, also promote RPE cell viability upon encapsulation (Parker et al., 2016). In addition, chitosan and alginate hydrogels with moduli similar to the retina have shown promise as substrates for differentiating and transplantion of RPCs and RPE, respectively (Hunt et al., 2017; Worthington et al., 2016a).
In addition to chemistry and mechanical behavior, the morphology of retinal biomaterials has also been a recent focus in the field. For example, the presence of randomly distributed pores reportedly maximizes RPC density on degradable polymer films, but the size of the pores does not significantly affect survival or differentiation (Calejo et al., 2017; Worthington et al., 2016b). It is clear that maximizing substrate surface area and facilitating nutrient diffusion throughout the scaffold are important design considerations. Furthermore, many (polymer-free) transplanted cells self-assemble into neural rosettes post-transplantation (Shirai et al., 2016). While this behavior is generally taken as a positive sign of differentiation to desired cell types, it reduces the ability of transplanted RPCs to integrate with host tissue and restore visual function. Thus, an ideal retinal cell transplantation material would possess, in addition to the features already discussed, the physical cues necessary to encourage RPC alignment (Figure 4). Creating tightly controlled polymer structures at relevant size scales has been historically difficult, but recent advances in 3D printing technologies have paved the way for the realization of this goal. In fact, the use of two-photon polymerization, a high-resolution 3D printing technique, can be used to create structures that facilitate RPC alignment (Worthington et al., 2017). We anticipate that future studies will focus on applying this technique to diverse chemistries and optimizing the resulting structures for use in the retina (Figure 4).
Figure 4. Human retinal tissue engineering.
The inner retina, which is preserved during inherited retinal degeneration, and the photoreceptor cell layer are intimately connected (schematic shown in A). The outer nuclear layer (ONL) comprises photoreceptor cell bodies stacked in columns while the outer segments are tightly packed side-by-side in close contact with the RPE (not shown). When viewed en face via phase contrast microscopy (B), human photoreceptor cells appear very tightly packed in a hexagonal array. This architecture can be closely recapitulated using high-resolution 3D printing (C-E), enabling the creation of photoreceptor cell scaffolds (F-H) meant to enable proper function and integration with the host inner retina. These highly tunable structures can facilitate close cell packing, guide cell orientation, and release small molecules that encourage synaptogenesis (I). GCL – ganglion cell layer, IPL – inner plexiform layer, INL – inner nuclear layer, OPL – outer plexiform layer, ONL – outer nuclear layer, IS/OS – photoreceptor cell inner and outer segments.
Reactive gliosis is known to be a significant barrier to neural regeneration in the central nervous system, including the retina (Gonzalez-Cordero et al., 2013; Tucker et al., 2010; Yiu and He, 2006; Y. Zhang et al., 2007). Delivery of specific small molecules or proteins can mitigate this phenomenon while also encouraging synaptogenesis (J. Ma et al., 2011; Tucker et al., 2010; Yao et al., 2011). However, precise spatial and temporal control of dosing is most likely to be achieved using polymeric delivery devices (Marquardt et al., 2015). In the past several years, groups have demonstrated that by embedding various combinations of stem-cell derived neurons, neurotrophic factors, axonal growth inhibitors and anti-gliotic enzymes in polymer scaffolds improves functional outcomes in models of spinal cord injury (Elliott Donaghue et al., 2016; 2015; Führmann et al., 2015; Wilems et al., 2015; Wilems and Sakiyama-Elbert, 2015), traumatic brain injury (X. Li et al., 2016) and stroke (Moshayedi et al., 2016; Nih et al., 2017). Many of these strategies could be implemented in retinal tissue engineering, with temporal release of such molecules being a “fourth dimension.”
There is also growing evidence to suggest that simultaneous transplantation of multiple cell types will be beneficial or even necessary for functional restoration in the retina. Transplantation of a sheet of autologous RPE alone in late-stage AMD patients was found to be well tolerated out to at least one year, which is an extraordinary and hopeful finding in itself (Mandai et al., 2017). However, functional restoration is not likely to be achieved without the delivery of replacement photoreceptor cells. Similarly, in cases such as this where both RPE and photoreceptor cells have degenerated, photoreceptor cells alone may not thrive without co-transplantation of RPE. This phenomenon is further complicated by the role of the choroidal vasculature in AMD disease progression. It too may need to be replaced or encouraged to regenerate in some cases in order to achieve restoration of proper retinal function. In addition to existing protocols for RPE and RPC differentiation, the recent development of SC-derived choroidal endothelial cell differentiation techniques will enable these multilayered interactions to be evaluated in vitro and in vivo (Chirco et al., 2017; Songstad et al., 2017; 2015).
8. Summary and Conclusions
Since the initial studies reporting the use of the CRISPR-Cas9 system for mammalian genome editing were published, the technology has been used extensively throughout the world for applications ranging from the creation of animal models to in vivo gene correction. Unlike its predecessors, TALENs and ZFNs, the CRISPR system is fairly simple, requiring nothing more than a basic knowledge of molecular biology to be able to effectively adopt the technology. As summarized in this review, one of the greatest areas of interest has been in vivo gene correction. In tissues such as the liver, which contains mitotically active cells, in vivo genome editing has been highly efficient. Unfortunately, as we have shown in a large animal model of retinal degeneration (Burnight et al., 2017), photoreceptor cell targeting and efficiency of NHEJ-based mutant gene deletion is currently so low that it would not be expected to provide a therapeutic benefit. The efficiency of HDR-based editing, which will be required to correct the majority of retinal disease-causing mutations (Stone et al., 2017), is expected to be even lower, and it is therefore difficult to imagine that this technology will take the place of traditional gene augmentation for the treatment of early stage retinal degeneration anytime soon. However, several groups around the world are aggressively pursuing ways to increase editing efficiency while simultaneously decreasing off target events.
Although the efficiency of targeted gene editing in mitotically active cells such as those found in the liver is significantly higher than in post-mitotic retinal neurons, so too are the risks of having an off-target event cause an untoward serious adverse reaction. As indicated above, the eye is unique in that it can be closely monitored following treatment using standard clinical approaches. If a CRISPR-induced significant adverse event were to occur, it could be readily detected and treated with noninvasive laser photocoagulation to prevent escape of mutant cells from the eye. Ocular diseases that can be treated by restoring function to a relatively small number of host cells may be the ideal targets for trials of in vivo CRISPR-based therapy in the near term. For instance, the trabecular meshwork (TM) is a tissue located at the junction of the cornea and the sclera that is involved in the maintenance of intraocular pressure via its resistance to drainage of aqueous humor from the eye. Genetic alteration of a relatively small proportion of trabecular meshwork cells could have a significant effect on the intraocular pressure just as laser photocoagulation of a discrete portion of the meshwork has been shown to do (Stein and Challa, 2007). For example, a recent study by Jain and colleagues demonstrated that CRISPR-mediated deletion of mutant myocilin in mouse TM cells in vivo convincingly prevented mutation induced ER-stress, TM cell death, elevation of intraocular pressure and death of retinal ganglion cells (Jain et al., 2017).
One therapeutic use of the CRISPR-Cas9 system is virtually unaffected by its meager molecular efficiency: the correction of disease-causing mutations in patient-derived iPSCs for autologous cell replacement. Unlike in vivo genome editing, genetically corrected iPSCs can have their genomes thoroughly analyzed both before and after CRISPR correction using any one of the genome sequencing approaches described in section 2.7 above. Corrected clones that are found to be free of deleterious off target cleavage events can be subsequently expanded and fully validated prior to differentiation. For photoreceptor cell replacement, clonally expanded CRISPR-corrected iPSC lines determined to be the most efficient producers of photoreceptor cells can subsequently be used to generate retinal cell grafts for autologous treatment of end stage retinal degenerative blindness.
As with any powerful technology, the CRISPR-Cas 9 system can be misused in ways that could jeopardize its use to make medically meaningful advancements. For example, if an investigator performs experiments that are perceived by the public to be unethical, it could have a very negative impact on the use of the technology for other purposes. Recently, CRISPR-based editing of human embryos was reported. The authors employed allele-specific sgRNAs and a S. pyogenes Cas9 along with ssODNs to correct a heterozygous variant in the MYBPC3 gene (H. Ma et al., 2017), mutations in which cause hypertrophic cardiomyopathy (Carrier et al., 2015). These embryos developed normally to blastocyst and ES cells without cytogenetic abnormalities, and whole genome sequencing and Digenome-seq evaluation of off-target cleavage did not reveal any detectable off-target mutations (H. Ma et al., 2017). The argument made by the authors of this study was that by using the CRISPR system it would be possible for individuals who were at risk of having a child affected with a potentially fatal disease to have unaffected children. The desire to have a child free from a known lethal genetic disorder is very understandable. This is the rationale behind the well-established method of preimplantation genetic testing, which unlike the CRISPR system does not have a risk of inducing off target modifications with severe developmental implications. The question that physicians, scientists and ethicists must constantly ask is whether the risks associated with an experiment are greater than the potential benefit. It is the opinion of these authors that CRISPR-based genome editing is very well suited and enormously valuable for genetic correction of disease-causing mutations in patient-derived iPSCs in vitro, but should not be used to genetically manipulate human embryos.
Since the original reports describing the use of the CRISPR technology for genome editing, many studies have been published that demonstrate the utility of this system for applications ranging from in vitro genome editing to the generation of knock out animal models of disease. Although the holy grail of this technology is safe and efficient in vivo gene correction, significant additional scientific work will be needed before this technology can be used to treat post-mitotic tissues like the human retina. In its current form, CRISPR-based genome editing holds the greatest promise for its ability to genetically correct patient-derived stem cells. By transiently expressing CRISPR reagents in patient specific iPSCs in vitro, and performing whole-genome sequencing on a number of the resulting clones, one can readily generate autologous cell lines with no deleterious off target events. These genetically corrected cell lines can in turn be used to generate photoreceptor cells, which when loaded onto biocompatible cell delivery scaffolds, can be transplanted into the subretinal space for autologous photoreceptor cell replacement.
List of Abbreviations
- AAV
adeno-associated virus
- iPSC
induced pluripotent stem cell
- CRISPRs
clustered regularly interspaced short palindromic repeats
- SRSRs
short regularly spaced repeats
- ZFN
zinc finger nuclease
- TALEN
transcription activator-like effector nuclease
- crRNA
CRISPR-RNA
- Cas
CRISPR-associated
- tracrRNA
trans-activating crRNA
- sgRNA
single guide RNA
- PAM
protospacer adjacent motif
- DSB
double-strand break
- NHEJ
non-homologous end-joining
- SSBR
single-stranded base repair
- HDR
homology-directed repair
- SpCas9
S. pyogenes Cas9
- SaCas9
S. aureus Cas9
- StCas9
S. thermophilus Cas9
- NmCas9
N. meningitidis Cas9
- FnCas9
F. novicida Cas9
- CjCas9
C. jejuni Cas9
- GUIDE-seq
genome-wide, unbiased identification of DSBs enabled by sequencing
- NGS
next generation sequencing
- BLESS
breaks labeling, enrichment on streptavidin, and next-generations sequencing
- BLISS
Breaks Labeling In Situ and Sequencing
- AsCpfI
Acidaminococcus sp. CpfI
- LbCpfI
L. bacterium CpfI
- WGS
whole genome sequencing
- CIRCLE-seq
circularization for in vitro reporting of cleavage effects by sequencing
- ssODN
single-stranded oligodeoxynucleotide
- AMD
age-related macular degeneration
- CNV
choroidal neovascularization
- SNV
single-nucleotide variant
- PCR
polymerase chain reaction
- RFLP
restriction fragment length polymorphism
- CECs
choroidal endothelial cells
- RPE
retinal pigment epithelium
- SNP
single nucleotide polymorphism
- hRPCs
human retinal progenitor cells
- ESC
embryonic stem cell
- FACS
fluorescence activated cell sorting
- PCL
poly(caprolactone)
- PLA
poly(lactic acid)
- HA
hyaluronic acid
- A
adenosine
- G
guanine
- C
cytosine
- T
thymidine
- R
adenosine or guanine
- V
adenosine, guanine, or cytosine
- W
adenosine or thymidine
9. References
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F, 2016. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. doi: 10.1126/science.aaf5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adijanto J, Naash MI, 2015. Nanoparticle-based technologies for retinal gene therapy. Eur J Pharm Biopharm 95, 353–367. doi: 10.1016/j.ejpb.2014.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afione SA, Conrad CK, Kearns WG, Chunduru S, Adams R, Reynolds TC, Guggino WB, Cutting GR, Carter BJ, Flotte TR, 1996. In vivo model of adeno-associated virus vector persistence and rescue. J. Virol. 70, 3235–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati J, Atkinson JP, Gelfand BD, 2013. Immunology of age-related macular degeneration. Nature Reviews Immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Ambati BK, van Rooijen N, Ambati J, 2003. Macrophage depletion inhibits experimental choroidal neovascularization. … ophthalmology & visual …. [DOI] [PubMed] [Google Scholar]
- Anders C, Bargsten K, Jinek M, 2016. Structural Plasticity of PAM Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9. Mol. Cell 61, 895–902. doi: 10.1016/j.molcel.2016.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M, 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573. doi: 10.1038/nature13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari AM, Ahmed AK, Matsangos AE, Lay F, Born LJ, Marti G, Harmon JW, Sun Z, 2016. Cellular GFP Toxicity and Immunogenicity: Potential Confounders in in Vivo Cell Tracking Experiments. Stem Cell Rev 12, 553–559. doi: 10.1007/s12015-016-9670-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auricchio A, Kobinger G, Anand V, Hildinger M, O’Connor E, Maguire AM, Wilson JM, Bennett J, 2001. Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum. Mol. Genet. 10, 3075–3081. [DOI] [PubMed] [Google Scholar]
- Bainbridge JWB, Mehat MS, Sundaram V, Robbie SJ, Barker SE, Ripamonti C, Georgiadis A, Mowat FM, Beattie SG, Gardner PJ, Feathers KL, Luong VA, Yzer S, Balaggan K, Viswanathan A, de Ravel TJL, Casteels I, Holder GE, Tyler N, Fitzke FW, Weleber RG, Nardini M, Moore AT, Thompson DA, Petersen-Jones SM, Michaelides M, van den Born LI, Stockman A, Smith AJ, Rubin G, Ali RR, 2015. Long-term effect of gene therapy on Leber’s congenital amaurosis. N. Engl. J. Med. 372, 1887–1897. doi: 10.1056/NEJMoa1414221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge JWB, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N, Petersen-Jones S, Bhattacharya SS, Thrasher AJ, Fitzke FW, Carter BJ, Rubin GS, Moore AT, Ali RR, 2008. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 358, 2231–2239. doi: 10.1056/NEJMoa0802268 [DOI] [PubMed] [Google Scholar]
- Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, Levy R, Akhtar AA, Breunig JJ, Svendsen CN, Wang S, 2016. In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol. Ther. 24, 556–563. doi: 10.1038/mt.2015.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballios BG, Cooke MJ, Donaldson L, Coles BLK, Morshead CM, van der Kooy D, Shoichet MS, 2015. A Hyaluronan-Based Injectable Hydrogel Improves the Survival and Integration of Stem Cell Progeny following Transplantation. Stem Cell Reports 4, 1031–1045. doi: 10.1016/j.stemcr.2015.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea-Cramer AO, Wang W, Lu S-J, Singh MS, Luo C, Huo H, McClements ME, Barnard AR, MacLaren RE, Lanza R, 2016. Function of human pluripotent stem cell-derived photoreceptor progenitors in blind mice. Sci Rep 6, 29784. doi: 10.1038/srep29784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, van der Oost J, 2015. Bacteriophage exclusion, a new defense system. The EMBO Journal 34, 134–135. doi: 10.15252/embj.201490620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R, 2011. CRISPR-Cas Systems in Bacteria and Archaea: Versatile Small RNAs for Adaptive Defense and Regulation. 10.1146/annurev-genet-110410–132430 45, 273–297. doi: [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF, 2011. TAL Effectors: Customizable Proteins for DNA Targeting. Science 333, 1843–1846. doi: 10.1126/science.1204094 [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD, 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading, Engl.) 151, 2551–2561. doi: 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- Brant Fernandes RA, Koss MJ, Falabella P, Stefanini FR, Maia M, Diniz B, Ribeiro R, Hu Y, Hinton D, Clegg DO, Chader G, Humayun MS, 2016. An Innovative Surgical Technique for Subretinal Transplantation of Human Embryonic Stem Cell-Derived Retinal Pigmented Epithelium in Yucatan Mini Pigs: Preliminary Results. Ophthalmic Surg Lasers Imaging Retina 47, 342–351. doi: 10.3928/23258160-20160324-07 [DOI] [PubMed] [Google Scholar]
- Brees C, Fransen M, 2014. A cost-effective approach to microporate mammalian cells with the Neon Transfection System. Anal. Biochem. 466, 49–50. doi: 10.1016/j.ab.2014.08.017 [DOI] [PubMed] [Google Scholar]
- Buchrieser J, James W, Moore MD, 2017. Human Induced Pluripotent Stem Cell-Derived Macrophages Share Ontogeny with MYB-Independent Tissue-Resident Macrophages. Stem Cell Reports 8, 334–345. doi: 10.1016/j.stemcr.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnight ER, Bohrer LR, Giacalone JC, Klaahsen DL, Daggett HT, East JS, Madumba RA, Worthington KS, Mullins RF, Stone EM, Tucker BA, Wiley LA, 2018. CRISPR-Cas9-Mediated Correction of the 1.02 kb Common Deletion in CLN3 in Induced Pluripotent Stem Cells from Patients with Batten Disease. The CRISPR Journal 1, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnight ER, Gupta M, Wiley LA, Anfinson KR, Tran A, Triboulet R, Hoffmann JM, Klaahsen DL, Andorf JL, Jiao C, Sohn EH, Adur MK, Ross JW, Mullins RF, Daley GQ, Schlaeger TM, Stone EM, Tucker BA, 2017. Using CRISPR-Cas9 to Generate Gene-Corrected Autologous iPSCs for the Treatment of Inherited Retinal Degeneration. Mol. Ther. doi: 10.1016/j.ymthe.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnight ER, Wiley LA, Drack AV, Braun TA, Anfinson KR, Kaalberg EE, Halder JA, Affatigato LM, Mullins RF, Stone EM, Tucker BA, 2014. CEP290 gene transfer rescues Leber congenital amaurosis cellular phenotype. Gene Ther. doi: 10.1038/gt.2014.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SM, Church GM, 2015. CRISPR‐Mediated Gene Targeting of Human Induced Pluripotent Stem Cells. John Wiley & Sons, Inc., Hoboken, NJ, USA. doi: 10.1002/9780470151808.sc05a08s35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SM, Mali P, Church GM, 2014. Genome editing in human stem cells. Meth. Enzymol. 546, 119–138. doi: 10.1016/B978-0-12-801185-0.00006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SM, Ortiz L, Mali P, Aach J, Church GM, 2015. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 43, e21–e21. doi: 10.1093/nar/gku1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calejo MT, Ilmarinen T, Vuorimaa-Laukkanen E, Talvitie E, Hakola HM, Skottman H, Kellomäki M, 2017. Langmuir-Schaefer film deposition onto honeycomb porous films for retinal tissue engineering. Acta Biomater 54, 138–149. doi: 10.1016/j.actbio.2017.02.035 [DOI] [PubMed] [Google Scholar]
- Carrier L, Mearini G, Stathopoulou K, Cuello F, 2015. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene 573, 188–197. doi: 10.1016/j.gene.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian-Serrano A, Davies B, 2017. CRISPR-Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools. Mamm. Genome 28, 247–261. doi: 10.1007/s00335-017-9697-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerbini T, Luo Y, Rao MS, Zou J, 2015. Transfection, selection, and colony-picking of human induced pluripotent stem cells TALEN-targeted with a GFP gene into the AAVS1 safe harbor. J Vis Exp. doi: 10.3791/52504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Hawes NL, Pardue MT, German AM, Hurd RE, Davisson MT, Nusinowitz S, Rengarajan K, Boyd AP, Sidney SS, Phillips MJ, Stewart RE, Chaudhury R, Nickerson JM, Heckenlively JR, Boatright JH, 2007. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision Res. 47, 624–633. doi: 10.1016/j.visres.2006.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JR, Lamba DA, Klesert TR, Torre AL, Hoshino A, Taylor RJ, Jayabalu A, Engel AL, Khuu TH, Wang RK, Neitz M, Neitz J, Reh TA, 2017. Transplantation of Human Embryonic Stem Cell-Derived Retinal Cells into the Subretinal Space of a Non-Human Primate. Transl Vis Sci Technol 6, 4–4. doi: 10.1167/tvst.6.3.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Ding X, Feng Y, Seebeck T, Jiang Y, Davis GD, 2017. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat Commun 8, 14958. doi: 10.1038/ncomms14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R, 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163–1171. doi: 10.1038/cr.2013.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery HR, McLenachan S, Humphries T, 2012. Accumulation of murine subretinal macrophages: effects of age, pigmentation and CX 3 CR1. Neurobiology of …. [DOI] [PubMed] [Google Scholar]
- Chirco KR, Worthington KS, Flamme-Wiese MJ, Riker MJ, Andrade JD, Ueberheide BM, Stone EM, Tucker BA, Mullins RF, 2017. Preparation and evaluation of human choroid extracellular matrix scaffolds for the study of cell replacement strategies. Acta Biomater 57, 293–303. doi: 10.1016/j.actbio.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV, Hauswirth WW, Aleman TS, Kaushal S, Schwartz SB, Boye SL, Windsor EAM, Conlon TJ, Sumaroka A, Pang J-J, Roman AJ, Byrne BJ, Jacobson SG, 2009. Human RPE65 gene therapy for Leber congenital amaurosis: persistence of early visual improvements and safety at 1 year. Hum. Gene Ther. 20, 999–1004. doi: 10.1089/hum.2009.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio A, Miselli F, Lombardo A, Marconi A, Malagoli Tagliazucchi G, Gonçalves MA, Pincelli C, Maruggi G, Del Rio M, Naldini L, Larcher F, Mavilio F, Recchia A, 2013. Targeted gene addition in human epithelial stem cells by zinc-finger nuclease-mediated homologous recombination. Mol. Ther. 21, 1695–1704. doi: 10.1038/mt.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Keebler JEM, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, Zilversmit M, Cartwright R, Rouleau GA, Daly M, Stone EA, Hurles ME, Awadalla P, 1000 Genomes Project, 2011. Variation in genome-wide mutation rates within and between human families. Nat. Genet. 43, 712–714. doi: 10.1038/ng.862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, Pasero P, Rowicka M, Dikic I, 2013. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365. doi: 10.1038/nmeth.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C, Corces VG, 2015. Topologically Associating Domains: An invariant framework or a dynamic scaffold? Nucleus 6, 430–434. doi: 10.1080/19491034.2015.1096467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Maizels N, 2014. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 111, E924–32. doi: 10.1073/pnas.1400236111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JY, Fan PD, Frizzell RA, 1996. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 7, 2101–2112. doi: 10.1089/hum.1996.7.17-2101 [DOI] [PubMed] [Google Scholar]
- Du D, Qi LS, 2016. CRISPR Technology for Genome Activation and Repression in Mammalian Cells. Cold Spring Harb Protoc 2016, pdb.prot090175. doi: 10.1101/pdb.prot090175 [DOI] [PubMed] [Google Scholar]
- Elliott Donaghue I, Tator CH, Shoichet MS, 2016. Local Delivery of Neurotrophin-3 and Anti-NogoA Promotes Repair After Spinal Cord Injury. Tissue Eng Part A 22, 733–741. doi: 10.1089/ten.TEA.2015.0471 [DOI] [PubMed] [Google Scholar]
- Elliott Donaghue I, Tator CH, Shoichet MS, 2015. Sustained delivery of bioactive neurotrophin-3 to the injured spinal cord. Biomater Sci 3, 65–72. doi: 10.1039/c4bm00311j [DOI] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M, 1998. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 18, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzmann V, Faude F, Wiedemann P, Kohen L, 1998. Immunological problems of transplantation into the subretinal space. Acta Anat (Basel) 162, 178–183. [DOI] [PubMed] [Google Scholar]
- Erez N, Milyavsky M, Eilam R, Shats I, Goldfinger N, Rotter V, 2003. Expression of prolyl-hydroxylase-1 (PHD1/EGLN2) suppresses hypoxia inducible factor-1alpha activation and inhibits tumor growth. Cancer Res. 63, 8777–8783. [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM, 2013. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121. doi: 10.1038/nmeth.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E, 2016. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. doi: 10.1038/nature17945 [DOI] [PubMed] [Google Scholar]
- Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, Kornepati AVR, Sousa A, Collins MA, Jayaram H, Cullen BR, Bumcrot D, 2015. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 16, 257. doi: 10.1186/s13059-015-0817-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JNC, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, Kim IK, Cho D, Zack D, Souied E, Scholl HPN, Bala E, Lee KE, Hunter DJ, Sardell RJ, Mitchell P, Merriam JE, Cipriani V, Hoffman JD, Schick T, Lechanteur YTE, Guymer RH, Johnson MP, Jiang Y, Stanton CM, Buitendijk GHS, Zhan X, Kwong AM, Boleda A, Brooks M, Gieser L, Ratnapriya R, Branham KE, Foerster JR, Heckenlively JR, Othman MI, Vote BJ, Liang HH, Souzeau E, McAllister IL, Isaacs T, Hall J, Lake S, Mackey DA, Constable IJ, Craig JE, Kitchner TE, Yang Z, Su Z, Luo H, Chen D, Ouyang H, Flagg K, Lin D, Mao G, Ferreyra H, Stark K, Strachwitz, von CN, Wolf A, Brandl C, Rudolph G, Olden M, Morrison MA, Morgan DJ, Schu M, Ahn J, Silvestri G, Tsironi EE, Park KH, Farrer LA, Orlin A, Brucker A, Li M, Curcio CA, Mohand-Saïd S, Sahel J-A, Audo I, Benchaboune M, Cree AJ, Rennie CA, Goverdhan SV, Grunin M, Hagbi-Levi S, Campochiaro P, Katsanis N, Holz FG, Blond F, Blanché H, Deleuze J-F, Igo RP, Truitt B, Peachey NS, Meuer SM, Myers CE, Moore EL, Klein R, Hauser MA, Postel EA, Courtenay MD, Schwartz SG, Kovach JL, Scott WK, Liew G, Tan AG, Gopinath B, Merriam JC, Smith RT, Khan JC, Shahid H, Moore AT, McGrath JA, Laux R, Brantley MA, Agarwal A, Ersoy L, Caramoy A, Langmann T, Saksens NTM, de Jong EK, Hoyng CB, Cain MS, Richardson AJ, Martin TM, Blangero J, Weeks DE, Dhillon B, van Duijn CM, Doheny KF, Romm J, Klaver CCW, Hayward C, Gorin MB, Klein ML, Baird PN, Hollander, den AI, Fauser S, Yates JRW, Allikmets R, Wang JJ, Schaumberg DA, Klein BEK, Hagstrom SA, Chowers I, Lotery AJ, Léveillard T, Zhang K, Brilliant MH, Hewitt AW, Swaroop A, Chew EY, Pericak-Vance MA, DeAngelis M, Stambolian D, Haines JL, Iyengar SK, Weber BHF, Abecasis GR, Heid IM, 2016. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 48, 134–143. doi: 10.1038/ng.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK, 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284. doi: 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Führmann T, Obermeyer J, Tator CH, Shoichet MS, 2015. Click-crosslinked injectable hyaluronic acid hydrogel is safe and biocompatible in the intrathecal space for ultimate use in regenerative strategies of the injured spinal cord. Methods 84, 60–69. doi: 10.1016/j.ymeth.2015.03.023 [DOI] [PubMed] [Google Scholar]
- Gao M, Zhang S, Liu C, Qin Y, Archacki S, Jin L, Wang Y, Liu F, Chen J, Liu Y, Wang J, Huang M, Liao S, Tang Z, Guo AY, Jiang F, Liu M, 2016. Whole exome sequencing identifies a novel NRL mutation in a Chinese family with autosomal dominant retinitis pigmentosa. Mol. Vis. 22, 234–242. [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V, 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, E2579–86. doi: 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Siksnys V, 2013. RNA-dependent DNA endonuclease Cas9 of the CRISPR system: Holy Grail of genome editing? Trends Microbiol. 21, 562–567. doi: 10.1016/j.tim.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Ghazi NG, Abboud EB, Nowilaty SR, Alkuraya H, Alhommadi A, Cai H, Hou R, Deng W-T, Boye SL, Almaghamsi A, Saikhan Al, F., Al-Dhibi H, Birch D, Chung C, Colak D, LaVail MM, Vollrath D, Erger K, Wang W, Conlon T, Zhang K, Hauswirth W, Alkuraya FS, 2016. Treatment of retinitis pigmentosa due to MERTK mutations by ocular subretinal injection of adeno-associated virus gene vector: results of a phase I trial. Hum. Genet. 135, 327–343. doi: 10.1007/s00439-016-1637-y [DOI] [PubMed] [Google Scholar]
- Giacalone JC, Sharma TP, Burnight ER, Fingert JH, Mullins RF, Stone EM, Tucker BA, 2018. CRISPR-Cas9 Based Genome Editing of Human Induced Pluripotent Stem Cells. Current Protocols. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS, 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi: 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Cordero A, West EL, Pearson RA, Duran Y, Carvalho LS, Chu CJ, Naeem A, Blackford SJI, Georgiadis A, Lakowski J, Hubank M, Smith AJ, Bainbridge JWB, Sowden JC, Ali RR, 2013. Photoreceptor precursors derived from three-dimensional embryonic stem cell cultures integrate and mature within adult degenerate retina. Nat. Biotechnol. 31, 741–747. doi: 10.1038/nbt.2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung H-L, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, Lee J-H, Loh Y-H, Manos PD, Montserrat N, Panopoulos AD, Ruiz S, Wilbert ML, Yu J, Kirkness EF, Izpisua Belmonte JC, Rossi DJ, Thomson JA, Eggan K, Daley GQ, Goldstein LSB, Zhang K, 2011. Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63–67. doi: 10.1038/nature09805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, Samulski RJ, 2005. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 79, 9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B-W, Apicella M, Mills J, Fan J-M, Reeves DA, French D, Podsakoff GM, Bessler M, Mason PJ, 2015. Impaired Telomere Maintenance and Decreased Canonical WNT Signaling but Normal Ribosome Biogenesis in Induced Pluripotent Stem Cells from X-Linked Dyskeratosis Congenita Patients. PLoS ONE 10, e0127414. doi: 10.1371/journal.pone.0127414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Kalle, Von C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le Deist F, Fischer A, Cavazzana-Calvo M, 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419. doi: 10.1126/science.1088547 [DOI] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud J-B, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly J-S, Concordet J-P, 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. doi: 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong DT, McGee TL, Sandberg MA, Berson EL, Asselbergs FW, van der Harst P, De Vivo I, Dryja TP, 2009. Search for a correlation between telomere length and severity of retinitis pigmentosa due to the dominant rhodopsin Pro23His mutation. Mol. Vis. 15, 592–597. [PMC free article] [PubMed] [Google Scholar]
- Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L, Conlon TJ, Boye SL, Flotte TR, Byrne BJ, Jacobson SG, 2008. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum. Gene Ther. 19, 979–990. doi: 10.1089/hum.2008.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Boutros M, 2014. E-CRISP: fast CRISPR target site identification. Nat. Methods 11, 122–123. doi: 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- Heman-Ackah SM, Bassett AR, Wood MJA, 2016. Precision Modulation of Neurodegenerative Disease-Related Gene Expression in Human iPSC-Derived Neurons. Sci Rep 6, 28420. doi: 10.1038/srep28420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans PW, Van Soolingen D, Bik EM, de Haas PE, Dale JW, van Embden JD, 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59, 2695–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, Nishimasu H, Nureki O, 2016. Structure and Engineering of Francisella novicida Cas9. Cell 164, 950–961. doi: 10.1016/j.cell.2016.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Nishimasu H, Ishitani R, Nureki O, 2016. Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9. Mol. Cell 61, 886–894. doi: 10.1016/j.molcel.2016.02.018 [DOI] [PubMed] [Google Scholar]
- Holkers M, Cathomen T, Gonçalves MAFV, 2014. Construction and characterization of adenoviral vectors for the delivery of TALENs into human cells. Methods 69, 179–187. doi: 10.1016/j.ymeth.2014.02.017 [DOI] [PubMed] [Google Scholar]
- Hollander, den AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KEJ, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, Hoyng CB, van den Born LI, Rohrschneider K, Cremers FPM, 2006. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 79, 556–561. doi: 10.1086/507318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homayouni M, 2009. Vascular endothelial growth factors and their inhibitors in ocular neovascular disorders. J Ophthalmic Vis Res 4, 105–114. [PMC free article] [PubMed] [Google Scholar]
- Hong SG, Yada RC, Choi K, Carpentier A, Liang TJ, Merling RK, Sweeney CL, Malech HL, Jung M, Corat MAF, AlJanahi AA, Lin Y, Liu H, Tunc I, Wang X, Palisoc M, Pittaluga S, Boehm M, Winkler T, Zou J, Dunbar CE, 2017. Rhesus iPSC Safe Harbor Gene-Editing Platform for Stable Expression of Transgenes in Differentiated Cells of All Germ Layers. Mol. Ther. 25, 44–53. doi: 10.1016/j.ymthe.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coûté-Monvoisin A-C, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R, 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190, 1401–1412. doi: 10.1128/JB.01415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu L-F, Sontheimer EJ, Thomson JA, 2013. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. U.S.A. 110, 15644–15649. doi: 10.1073/pnas.1313587110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F, 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi: 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Xiaosong, Wang Y, Yan W, Smith C, Ye Z, Wang J, Gao Y, Mendelsohn L, Cheng L, 2015. Production of Gene-Corrected Adult Beta Globin Protein in Human Erythrocytes Differentiated from Patient iPSCs After Genome Editing of the Sickle Point Mutation. Stem Cells 33, 1470–1479. doi: 10.1002/stem.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Xionggao, Zhou G, Wu W, Duan Y, Ma G, Song J, Xiao R, Vandenberghe L, Zhang F, D’Amore PA, Lei H, 2017. Genome editing abrogates angiogenesis in vivo. Nat Commun 8, 112. doi: 10.1038/s41467-017-00140-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SSC, Chrysostomou V, Li F, Lim JKH, Wang J-H, Powell JE, Tu L, Daniszewski M, Lo C, Wong RC, Crowston JG, Pébay A, King AE, Bui BV, Liu G-S, Hewitt AW, 2016. AAV-Mediated CRISPR/Cas Gene Editing of Retinal Cells In Vivo. Invest. Ophthalmol. Vis. Sci. 57, 3470–3476. doi: 10.1167/iovs.16-19316 [DOI] [PubMed] [Google Scholar]
- Hunt NC, Hallam D, Karimi A, Mellough CB, Chen J, Steel DHW, Lako M, 2017. 3D culture of human pluripotent stem cells in RGD-alginate hydrogel improves retinal tissue development. Acta Biomater 49, 329–343. doi: 10.1016/j.actbio.2016.11.016 [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Kaur P, Smith PP, Perez-Reyes N, Blanton RA, McDougall JK, 1991. Progression of human papillomavirus type 18-immortalized human keratinocytes to a malignant phenotype. Proceedings of the National Academy of Sciences 88, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, Lundin K, Mikkola M, Trokovic R, Peitz M, Brüstle O, Bazett-Jones DP, Alitalo K, Lahesmaa R, Nagy A, Otonkoski T, 2011. Copy number variation and selection during reprogramming to pluripotency. Nature 471, 58–62. doi: 10.1038/nature09871 [DOI] [PubMed] [Google Scholar]
- Iijima S, 1991. Helical microtubules of graphitic carbon. Nature. [Google Scholar]
- Ilmarinen T, Hiidenmaa H, Kööbi P, Nymark S, Sorkio A, Wang J-H, Stanzel BV, Thieltges F, Alajuuma P, Oksala O, Kataja M, Uusitalo H, Skottman H, 2015. Ultrathin Polyimide Membrane as Cell Carrier for Subretinal Transplantation of Human Embryonic Stem Cell Derived Retinal Pigment Epithelium. PLoS ONE 10, e0143669. doi: 10.1371/journal.pone.0143669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A, 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu T, Higo S, Masumura Y, Kohama Y, Shiba M, Higo T, Shibamoto M, Nakagawa A, Morimoto S, Takashima S, Hikoso S, Sakata Y, 2017. Targeted Genome Replacement via Homology-directed Repair in Non-dividing Cardiomyocytes. Sci Rep 7, 9363. doi: 10.1038/s41598-017-09716-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyombe-Engembe J-P, Ouellet DL, Barbeau X, Rousseau J, Chapdelaine P, Lagüe P, Tremblay JP, 2016. Efficient Restoration of the Dystrophin Gene Reading Frame and Protein Structure in DMD Myoblasts Using the CinDel Method. Mol Ther Nucleic Acids 5, e283. doi: 10.1038/mtna.2015.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Ratnakaram R, Heon E, Schwartz SB, Roman AJ, Peden MC, Aleman TS, Boye SL, Sumaroka A, Conlon TJ, Calcedo R, Pang J-J, Erger KE, Olivares MB, Mullins CL, Swider M, Kaushal S, Feuer WJ, Iannaccone A, Fishman GA, Stone EM, Byrne BJ, Hauswirth WW, 2012. Gene therapy for leber congenital amaurosis caused by RPE65 mutations: safety and efficacy in 15 children and adults followed up to 3 years. Arch. Ophthalmol. 130, 9–24. doi: 10.1001/archophthalmol.2011.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Zode G, Kasetti RB, Ran FA, Yan W, Sharma TP, Bugge K, Searby CC, Fingert JH, Zhang F, Clark AF, Sheffield VC, 2017. CRISPR-Cas9-based treatment of myocilin-associated glaucoma. Proc. Natl. Acad. Sci. U.S.A. 114, 11199–11204. doi: 10.1073/pnas.1706193114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA, 2015. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481. doi: 10.1126/science.aab1452 [DOI] [PubMed] [Google Scholar]
- Jiang H, Pierce GF, Ozelo MC, de Paula EV, Vargas JA, Smith P, Sommer J, Luk A, Manno CS, High KA, Arruda VR, 2006. Evidence of multiyear factor IX expression by AAV-mediated gene transfer to skeletal muscle in an individual with severe hemophilia B. Mol. Ther. 14, 452–455. doi: 10.1016/j.ymthe.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J, 2013a. RNA-programmed genome editing in human cells. Elife 2, e00471. doi: 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J, 2013b. RNA-programmed genome editing in human cells. Elife. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA, 2014. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997–1247997. doi: 10.1126/science.1247997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay MA, Manno CS, Ragni MV, Larson PJ, Couto LB, McClelland A, Glader B, Chew AJ, Tai SJ, Herzog RW, Arruda V, Johnson F, Scallan C, Skarsgard E, Flake AW, High KA, 2000. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat. Genet. 24, 257–261. doi: 10.1038/73464 [DOI] [PubMed] [Google Scholar]
- Kearns NA, Genga RMJ, Enuameh MS, Garber M, Wolfe SA, Maehr R, 2014. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 141, 219–223. doi: 10.1242/dev.103341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim J-I, Kim J-S, 2015. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–43– 1 p following 243. doi: 10.1038/nmeth.3284 [DOI] [PubMed] [Google Scholar]
- Kim D, Kim S, Kim S, Park J, Kim J-S, 2016. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Research 26, 406–415. doi: 10.1101/gr.199588.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Koo T, Park SW, Kim D, Kim K, Cho H-Y, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim J-S, 2017. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8, 14500. doi: 10.1038/ncomms14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Park SW, Kim JH, Lee SH, Kim D, Koo T, Kim K-E, Kim JH, Kim J-S, 2017. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Research 27, 419–426. doi: 10.1101/gr.219089.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-T, Park J, Kim D, Kim K, Bae S, Schlesner M, Kim J-S, 2017. Questioning unexpected CRISPR off-target mutations in vivo. bioRxiv 157925. doi: 10.1101/157925 [DOI] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK, 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. doi: 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, Aryee MJ, Joung JK, 2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485. doi: 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koss MJ, Falabella P, Stefanini FR, Pfister M, Thomas BB, Kashani AH, Brant R, Zhu D, Clegg DO, Hinton DR, Humayun MS, 2016. Subretinal implantation of a monolayer of human embryonic stem cell-derived retinal pigment epithelium: a feasibility and safety study in Yucatán minipigs. Graefes Arch. Clin. Exp. Ophthalmol. 254, 1553–1565. doi: 10.1007/s00417-016-3386-y [DOI] [PubMed] [Google Scholar]
- Kundu J, Michaelson A, Baranov P, Chiumiento M, Nigl T, Young MJ, Carrier RL, 2017. Interphotoreceptor matrix based biomaterial: Impact on human retinal progenitor cell attachment and differentiation. J. Biomed. Mater. Res. Part B Appl. Biomater. 54, 1789. doi: 10.1002/jbm.b.33901 [DOI] [PubMed] [Google Scholar]
- Lakowski J, Gonzalez-Cordero A, West EL, Han Y-T, Welby E, Naeem A, Blackford SJI, Bainbridge JWB, Pearson RA, Ali RR, Sowden JC, 2015. Transplantation of Photoreceptor Precursors Isolated via a Cell Surface Biomarker Panel from Embryonic Stem Cell-Derived Self-Forming Retina. Stem Cells. doi: 10.1002/stem.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau C, Clement K, Hsu JY, Pattanayak V, Joung JK, Aryee MJ, Pinello L, 2017. “Unexpected mutations after CRISPR-Cas9 editing in vivo” are most likely pre-existing sequence variants and not nuclease-induced mutations. bioRxiv 159707. doi: 10.1101/159707 [DOI] [Google Scholar]
- Lawley E, Baranov P, Young M, 2015. Hybrid vitronectin-mimicking polycaprolactone scaffolds for human retinal progenitor cell differentiation and transplantation. J Biomater Appl 29, 894–902. doi: 10.1177/0885328214547751 [DOI] [PubMed] [Google Scholar]
- Leaderer D, Cashman SM, Kumar-Singh R, 2016. G-quartet oligonucleotide mediated delivery of proteins into photoreceptors and retinal pigment epithelium via intravitreal injection. Exp. Eye Res. 145, 380–392. doi: 10.1016/j.exer.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenay RT, Beisel CL, 2017. Deciphering, Communicating, and Engineering the CRISPR PAM. J. Mol. Biol. 429, 177–191. doi: 10.1016/j.jmb.2016.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolò M, Makino CL, Sidman RL, 1999. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proceedings of the National Academy of Sciences 96, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N, 1989. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 246, 1306–1309. [DOI] [PubMed] [Google Scholar]
- Li T, Snyder WK, Olsson JE, Dryja TP, 1996. Transgenic mice carrying the dominant rhodopsin mutation P347S: evidence for defective vectorial transport of rhodopsin to the outer segments. Proceedings of the National Academy of Sciences 93, 14176–14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tzeng SY, Liu X, Tammia M, Cheng Y-H, Rolfe A, Sun D, Zhang N, Green JJ, Wen X, Mao H-Q, 2016. Nanoparticle-mediated transcriptional modification enhances neuronal differentiation of human neural stem cells following transplantation in rat brain. Biomaterials 84, 157–166. doi: 10.1016/j.biomaterials.2016.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littink KW, Stappers PTY, Riemslag FCC, Talsma HE, van Genderen MM, Cremers FPM, Collin RWJ, van den Born LI, 2018. Autosomal Recessive NRL Mutations in Patients with Enhanced S-Cone Syndrome. Genes (Basel) 9, 68. doi: 10.3390/genes9020068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li X, Ma J, Li Z, You L, Wang J, Wang M, Zhang X, Wang Y, 2017. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 170, 714–726.e10. doi: 10.1016/j.cell.2017.06.050 [DOI] [PubMed] [Google Scholar]
- Lotery AJ, Yang GS, Mullins RF, Russell SR, Schmidt M, Stone EM, Lindbloom JD, Chiorini JA, Kotin RM, Davidson BL, 2003. Adeno-Associated Virus Type 5: Transduction Efficiency and Cell-Type Specificity in the Primate Retina. Hum. Gene Ther. 14, 1663–1671. doi: 10.1089/104303403322542301 [DOI] [PubMed] [Google Scholar]
- Luo Y, Xiao W, Zhu X, Mao Y, Liu X, Chen X, Huang J, Tang S, Rizzolo LJ, 2011. Differential expression of claudins in retinas during normal development and the angiogenesis of oxygen-induced retinopathy. Invest. Ophthalmol. Vis. Sci. 52, 7556–7564. doi: 10.1167/iovs.11-7185 [DOI] [PubMed] [Google Scholar]
- Ma H, Marti-Gutierrez N, Park S-W, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, Darby H, Van Dyken C, Li Y, Kang E, Park A-R, Kim D, Kim S-T, Gong J, Gu Y, Xu X, Battaglia D, Krieg SA, Lee DM, Wu DH, Wolf DP, Heitner SB, Belmonte JCI, Amato P, Kim J-S, Kaul S, Mitalipov S, 2017. Correction of a pathogenic gene mutation in human embryos. Nature 548, 413–419. doi: 10.1038/nature23305 [DOI] [PubMed] [Google Scholar]
- Ma J, Kabiel M, Tucker BA, Ge J, Young MJ, 2011. Combining chondroitinase ABC and growth factors promotes the integration of murine retinal progenitor cells transplanted into Rho(−/−) mice. Mol. Vis. 17, 1759–1770. [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK, 2013. CRISPR RNA–guided activation of endogenous human genes. Nat. Methods 10, 977–979. doi: 10.1038/nmeth.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio I, Holkers M, Liu J, Janssen JM, Chen X, Gonçalves MAFV, 2014. Adenoviral vector delivery of RNA-guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a diverse array of human cells. Sci Rep 4, 5105. doi: 10.1038/srep05105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, Mingozzi F, Bennicelli JL, Ying G-S, Rossi S, Fulton A, Marshall KA, Banfi S, Chung DC, Morgan JIW, Hauck B, Zelenaia O, Zhu X, Raffini L, Coppieters F, De Baere E, Shindler KS, Volpe NJ, Surace EM, Acerra C, Lyubarsky A, Redmond TM, Stone E, Sun J, McDonnell JW, Leroy BP, Simonelli F, Bennett J, 2009. Age-dependent effects of RPE65 gene therapy for Leber’s congenital amaurosis: a phase 1 dose-escalation trial. Lancet 374, 1597–1605. doi: 10.1016/S0140-6736(09)61836-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, Arruda VR, Konkle B, Stone E, Sun J, Jacobs J, Dell’Osso L, Hertle R, Ma J-X, Redmond TM, Zhu X, Hauck B, Zelenaia O, Shindler KS, Maguire MG, Wright JF, Volpe NJ, McDonnell JW, Auricchio A, High KA, Bennett J, 2008. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 358, 2240–2248. doi: 10.1056/NEJMoa0802315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV, 2011. Evolution and classification of the CRISPR-Cas systems. Nature Reviews. Microbiology 9, 467–477. doi: 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Güell M, DiCarlo JE, Norville JE, Church GM, 2013a. RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Güell M, DiCarlo JE, Norville JE, Church GM, 2013b. RNA-Guided Human Genome Engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai M, Watanabe A, Kurimoto Y, Hirami Y, Morinaga C, Daimon T, Fujihara M, Akimaru H, Sakai N, Shibata Y, Terada M, Nomiya Y, Tanishima S, Nakamura M, Kamao H, Sugita S, Onishi A, Ito T, Fujita K, Kawamata S, Go MJ, Shinohara C, Hata K-I, Sawada M, Yamamoto M, Ohta S, Ohara Y, Yoshida K, Kuwahara J, Kitano Y, Amano N, Umekage M, Kitaoka F, Tanaka A, Okada C, Takasu N, Ogawa S, Yamanaka S, Takahashi M, 2017. Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N. Engl. J. Med. 376, 1038–1046. doi: 10.1056/NEJMoa1608368 [DOI] [PubMed] [Google Scholar]
- Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, Judge LM, Gordon DE, Eskildsen TV, Villalta JE, Horlbeck MA, Gilbert LA, Krogan NJ, Sheikh SP, Weissman JS, Qi LS, So P-L, Conklin BR, 2016. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 18, 541–553. doi: 10.1016/j.stem.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt LM, Ee X, Iyer N, Hunter D, Mackinnon SE, Wood MD, Sakiyama-Elbert SE, 2015. Finely Tuned Temporal and Spatial Delivery of GDNF Promotes Enhanced Nerve Regeneration in a Long Nerve Defect Model. Tissue Eng Part A 21, 2852–2864. doi: 10.1089/ten.TEA.2015.0311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill TJ, Bohana-Kashtan O, Stoddard JW, Andrews MD, Pandit N, Rosenberg-Belmaker LR, Wiser O, Matzrafi L, Banin E, Reubinoff B, Netzer N, Irving C, 2017. Long-Term Efficacy of GMP Grade Xeno-Free hESC-Derived RPE Cells Following Transplantation. Transl Vis Sci Technol 6, 17–17. doi: 10.1167/tvst.6.3.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill TJ, Prusky GT, Luna G, LaVail MM, Fisher SK, Lewis GP, 2012. Optomotor and immunohistochemical changes in the juvenile S334ter rat. Exp. Eye Res. 104, 65–73. doi: 10.1016/j.exer.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier ID, Bernreuther C, Tilling T, Neidhardt J, Wong YW, Schulze C, Streichert T, Schachner M, 2010. Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J. 24, 1714–1724. doi: 10.1096/fj.09-140749 [DOI] [PubMed] [Google Scholar]
- Mendell JR, Sahenk Z, Malik V, Gomez AM, Flanigan KM, Lowes LP, Alfano LN, Berry K, Meadows E, Lewis S, Braun L, Shontz K, Rouhana M, Clark KR, Rosales XQ, Al-Zaidy S, Govoni A, Rodino-Klapac LR, Hogan MJ, Kaspar BK, 2015. A phase 1/2a follistatin gene therapy trial for becker muscular dystrophy. Mol. Ther. 23, 192–201. doi: 10.1038/mt.2014.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Neuhausser WM, Santos D, Valen E, Gagnon JA, Maas K, Sandoe J, Schier AF, Eggan K, 2015. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep 11, 875–883. doi: 10.1016/j.celrep.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitzelfelt KA, McDermott-Roe C, Grzybowski MN, Marquez M, Kuo C-T, Riedel M, Lai S, Choi MJ, Kolander KD, Helbling D, Dimmock DP, Battle MA, Jou CJ, Tristani-Firouzi M, Verbsky JW, Benjamin IJ, Geurts AM, 2017. Efficient Precision Genome Editing in iPSCs via Genetic Co-targeting with Selection. Stem Cell Reports 8, 491–499. doi: 10.1016/j.stemcr.2017.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Berman JR, Cooper SB, Mayerl SJ, Chan AH, Zhang B, Karlin-Neumann GA, Conklin BR, 2016. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci Rep 6, 23549. doi: 10.1038/srep23549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr JC, de Pablo JJ, Palecek SP, 2006. Electroporation of human embryonic stem cells: Small and macromolecule loading and DNA transfection. Biotechnol. Prog. 22, 825–834. doi: 10.1021/bp0600334 [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E, 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- Monteys AM, Ebanks SA, Keiser MS, Davidson BL, 2017. CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo. Mol. Ther. 25, 12–23. doi: 10.1016/j.ymthe.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshayedi P, Nih LR, Llorente IL, Berg AR, Cinkornpumin J, Lowry WE, Segura T, Carmichael ST, 2016. Systematic optimization of an engineered hydrogel allows for selective control of human neural stem cell survival and differentiation after transplantation in the stroke brain. Biomaterials 105, 145–155. doi: 10.1016/j.biomaterials.2016.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C, Gernoux G, Gruntman AM, Borel F, Reeves EP, Calcedo R, Rouhani FN, Yachnis A, Humphries M, Campbell-Thompson M, Messina L, Chulay JD, Trapnell B, Wilson JM, McElvaney NG, Flotte TR, 2017. 5 Year Expression and Neutrophil Defect Repair after Gene Therapy in Alpha-1 Antitrypsin Deficiency. Mol. Ther. 25, 1387–1394. doi: 10.1016/j.ymthe.2017.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RF, Kuehn MH, Faidley EA, Syed NA, Stone EM, 2007. Differential macular and peripheral expression of bestrophin in human eyes and its implication for best disease. Invest. Ophthalmol. Vis. Sci. 48, 3372–3380. doi: 10.1167/iovs.06-0868 [DOI] [PubMed] [Google Scholar]
- Mullins RF, Kuehn MH, Radu RA, Enriquez GS, East JS, Schindler EI, Travis GH, Stone EM, 2012. Autosomal recessive retinitis pigmentosa due to ABCA4 mutations: clinical, pathologic, and molecular characterization. Invest. Ophthalmol. Vis. Sci. 53, 1883. doi: 10.1167/iovs.12-9477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C, 2016. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol. Ther. 24, 636–644. doi: 10.1038/mt.2015.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata A, Amemura M, Makino K, 1989. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J. Bacteriol. 171, 3553–3556. doi: 10.1128/jb.171.6.3553-3556.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S, Herzog RW, 2010. Progress and prospects: immune responses to viral vectors. Gene Ther. 17, 295–304. doi: 10.1038/gt.2009.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nih LR, Moshayedi P, Llorente IL, Berg AR, Cinkornpumin J, Lowry WE, Segura T, Carmichael ST, 2017. Engineered HA hydrogel for stem cell transplantation in the brain: Biocompatibility data using a design of experiment approach. Data Brief 10, 202–209. doi: 10.1016/j.dib.2016.11.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O, 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. doi: 10.1016/j.cell.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nommiste B, Fynes K, Tovell VE, Ramsden C, da Cruz L, Coffey P, 2017. Stem cell-derived retinal pigment epithelium transplantation for treatment of retinal disease. Prog. Brain Res. 231, 225–244. doi: 10.1016/bs.pbr.2017.03.003 [DOI] [PubMed] [Google Scholar]
- Ochakovski GA, Bartz-Schmidt KU, Fischer MD, 2017. Retinal Gene Therapy: Surgical Vector Delivery in the Translation to Clinical Trials. Front Neurosci 11, 174. doi: 10.3389/fnins.2017.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP, 1992. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron 9, 815–830. [DOI] [PubMed] [Google Scholar]
- Ouellet DL, Cherif K, Rousseau J, Tremblay JP, 2017. Deletion of the GAA repeats from the human frataxin gene using the CRISPR-Cas9 system in YG8R-derived cells and mouse models of Friedreich ataxia. Gene Ther. 24, 265–274. doi: 10.1038/gt.2016.89 [DOI] [PubMed] [Google Scholar]
- Park I-H, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ, 2008. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146. doi: 10.1038/nature06534 [DOI] [PubMed] [Google Scholar]
- Parker J, Mitrousis N, Shoichet MS, 2016. Hydrogel for Simultaneous Tunable Growth Factor Delivery and Enhanced Viability of Encapsulated Cells in Vitro. Biomacromolecules 17, 476–484. doi: 10.1021/acs.biomac.5b01366 [DOI] [PubMed] [Google Scholar]
- Pennington KL, Deangelis MM, 2016. Epidemiology of age-related macular degeneration (AMD): associations with cardiovascular disease phenotypes and lipid factors. Eye Vis (Lond) 3, 34. doi: 10.1186/s40662-016-0063-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Delphin N, Hanein S, Gerber S, Dufier J-L, Roche O, Defoort-Dhellemmes S, Dollfus H, Fazzi E, Munnich A, Kaplan J, Rozet J-M, 2007. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum. Mutat. 28, 416–416. doi: 10.1002/humu.9485 [DOI] [PubMed] [Google Scholar]
- Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ, 1996. Long-range disruption of gene expression by a selectable marker cassette. Proceedings of the National Academy of Sciences 93, 13090–13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza Reyes A, Petrus-Reurer S, Antonsson L, Stenfelt S, Bartuma H, Panula S, Mader T, Douagi I, André H, Hovatta O, Lanner F, Kvanta A, 2016. Xeno-Free and Defined Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells Functionally Integrate in a Large-Eyed Preclinical Model. Stem Cell Reports 6, 9–17. doi: 10.1016/j.stemcr.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G, 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663. doi: 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F, 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. doi: 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F, 2013a. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389. doi: 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013b. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. doi: 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refaey El, M., Xu L, Gao Y, Canan BD, Adesanya TA, Warner SC, Akagi K, Symer DE, Mohler PJ, Ma J, Janssen PM, Han R, 2017. In Vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice. Circ. Res. CIRCRESAHA.117.310996. doi: 10.1161/CIRCRESAHA.117.310996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera M, Fontrodona L, Albert S, Ramirez DM, Seriola A, Salas A, Muñoz Y, Ramos D, Villegas-Perez MP, Zapata MA, Raya A, Ruberte J, Veiga A, Garcia-Arumi J, 2016. Comparative study of human embryonic stem cells (hESC) and human induced pluripotent stem cells (hiPSC) as a treatment for retinal dystrophies. Mol Ther Methods Clin Dev 3, 16010. doi: 10.1038/mtm.2016.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MK, Vermerris W, 2017. Recent Advances in Nanomaterials for Gene Delivery-A Review. Nanomaterials (Basel) 7, 94. doi: 10.3390/nano7050094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière I, Dunbar CE, Sadelain M, 2012. Hematopoietic stem cell engineering at a crossroads. Blood 119, 1107–1116. doi: 10.1182/blood-2011-09-349993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan G-X, Barry E, Yu D, Lukason M, Cheng SH, Scaria A, 2017. CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol. Ther. 25, 331–341. doi: 10.1016/j.ymthe.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S, Bennett J, Wellman JA, Chung DC, Yu Z-F, Tillman A, Wittes J, Pappas J, Elci O, McCague S, Cross D, Marshall KA, Walshire J, Kehoe TL, Reichert H, Davis M, Raffini L, George LA, Hudson FP, Dingfield L, Zhu X, Haller JA, Sohn EH, Mahajan VB, Pfeifer W, Weckmann M, Johnson C, Gewaily D, Drack A, Stone E, Wachtel K, Simonelli F, Leroy BP, Wright JF, High KA, Maguire AM, 2017. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. doi: 10.1016/S0140-6736(17)31868-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutar M, Provis JM, Valter K, 2010. Brief exposure to damaging light causes focal recruitment of macrophages, and long-term destabilization of photoreceptors in the albino rat retina. Curr. Eye Res. [DOI] [PubMed] [Google Scholar]
- Sadelain M, Papapetrou EP, Bushman FD, 2011. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer 12, 51–58. doi: 10.1038/nrc3179 [DOI] [PubMed] [Google Scholar]
- Sampson TR, Weiss DS, 2013. Cas9-dependent endogenous gene regulation is required for bacterial virulence. Biochem. Soc. Trans. 41, 1407–1411. doi: 10.1042/BST20130163 [DOI] [PubMed] [Google Scholar]
- Sander JD, Joung JK, 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355. doi: 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Ferreira T, Völkner M, Borsch O, Haas J, Cimalla P, Vasudevan P, Carmeliet P, Corbeil D, Michalakis S, Koch E, Karl MO, Ader M, 2016. Stem Cell-Derived Photoreceptor Transplants Differentially Integrate Into Mouse Models of Cone-Rod Dystrophy. Invest. Ophthalmol. Vis. Sci. 57, 3509–3520. doi: 10.1167/iovs.16-19087 [DOI] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V, 2011. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282. doi: 10.1093/nar/gkr606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer KA, Wu W-H, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB, 2017. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat. Methods 14, 547–548. doi: 10.1038/nmeth.4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Erfurth U, Kaiser PK, Korobelnik J-F, Brown DM, Chong V, Nguyen QD, Ho AC, Ogura Y, Simader C, Jaffe GJ, Slakter JS, Yancopoulos GD, Stahl N, Vitti R, Berliner AJ, Soo Y, Anderesi M, Sowade O, Zeitz O, Norenberg C, Sandbrink R, Heier JS, 2014. Intravitreal aflibercept injection for neovascular age-related macular degeneration: ninety-six-week results of the VIEW studies. Ophthalmology 121, 193–201. doi: 10.1016/j.ophtha.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Schwartz SD, Regillo CD, Lam BL, Eliott D, Rosenfeld PJ, Gregori NZ, Hubschman J-P, Davis JL, Heilwell G, Spirn M, Maguire J, Gay R, Bateman J, Ostrick RM, Morris D, Vincent M, Anglade E, Del Priore LV, Lanza R, 2015. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet 385, 509–516. doi: 10.1016/S0140-6736(14)61376-3 [DOI] [PubMed] [Google Scholar]
- Seo S, Mullins RF, Dumitrescu AV, Bhattarai S, Gratie D, Wang K, Stone EM, Sheffield V, Drack AV, 2013. Subretinal gene therapy of mice with bardet-biedl syndrome type 1. Invest. Ophthalmol. Vis. Sci. 54, 6118–6132. doi: 10.1167/iovs.13-11673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Zhou P-Y, Peng G-H, 2017. Experimental Study of the Biological Properties of Human Embryonic Stem Cell-Derived Retinal Progenitor Cells. Sci Rep 7, 42363. doi: 10.1038/srep42363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield VC, Stone EM, 2011. Genomics and the eye. N. Engl. J. Med. 364, 1932–42. [DOI] [PubMed] [Google Scholar]
- Shin JW, Kim K-H, Chao MJ, Atwal RS, Gillis T, MacDonald ME, Gusella JF, Lee J-M, 2016. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 25, 4566–4576. doi: 10.1093/hmg/ddw286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai H, Mandai M, Matsushita K, Kuwahara A, Yonemura S, Nakano T, Assawachananont J, Kimura T, Saito K, Terasaki H, Eiraku M, Sasai Y, Takahashi M, 2016. Transplantation of human embryonic stem cell-derived retinal tissue in two primate models of retinal degeneration. Proc. Natl. Acad. Sci. U.S.A. 113, E81–90. doi: 10.1073/pnas.1512590113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV, 2015. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 60, 385–397. doi: 10.1016/j.molcel.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL, Rossi S, Marshall K, Banfi S, Surace EM, Sun J, Redmond TM, Zhu X, Shindler KS, Ying G-S, Ziviello C, Acerra C, Wright JF, McDonnell JW, High KA, Bennett J, Auricchio A, 2010. Gene therapy for Leber’s congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 18, 643–650. doi: 10.1038/mt.2009.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MS, Charbel Issa P, Butler R, Martin C, Lipinski DM, Sekaran S, Barnard AR, MacLaren RE, 2013. Reversal of end-stage retinal degeneration and restoration of visual function by photoreceptor transplantation. Proc. Natl. Acad. Sci. U.S.A. 110, 1101–1106. doi: 10.1073/pnas.1119416110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V, 2011. Cas3 is a single‐stranded DNA nuclease and ATP‐dependent helicase in the CRISPR/Cas immune system. The EMBO Journal 30, 1335–1342. doi: 10.1038/emboj.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BK, Martin AD, Lawson LA, Vernot V, Marcus J, Islam S, Shafi N, Corti M, Collins SW, Byrne BJ, 2017. Inspiratory muscle conditioning exercise and diaphragm gene therapy in Pompe disease: Clinical evidence of respiratory plasticity. Exp. Neurol. 287, 216–224. doi: 10.1016/j.expneurol.2016.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WK, Park K-M, Kim H-J, Lee JH, Choi J, Chong SY, Shim SH, Del Priore LV, Lanza R, 2015. Treatment of macular degeneration using embryonic stem cell-derived retinal pigment epithelium: preliminary results in Asian patients. Stem Cell Reports 4, 860–872. doi: 10.1016/j.stemcr.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songstad AE, Wiley LA, Duong K, Kaalberg E, Flamme-Wiese MJ, Cranston CM, Riker MJ, Levasseur D, Stone EM, Mullins RF, Tucker BA, 2015. Generating iPSC-Derived Choroidal Endothelial Cells to Study Age-Related Macular Degeneration. Invest. Ophthalmol. Vis. Sci. 56, 8258–8267. doi: 10.1167/iovs.15-17073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songstad AE, Worthington KS, Chirco KR, Giacalone JC, Whitmore SS, Anfinson KR, Ochoa D, Cranston CM, Riker MJ, Neiman M, Stone EM, Mullins RF, Tucker BA, 2017. Connective Tissue Growth Factor Promotes Efficient Generation of Human Induced Pluripotent Stem Cell-Derived Choroidal Endothelium. Stem Cells Transl Med 6, 1533–1546. doi: 10.1002/sctm.16-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkio A, Porter PJ, Juuti-Uusitalo K, Meenan BJ, Skottman H, Burke GA, 2015. Surface Modified Biodegradable Electrospun Membranes as a Carrier for Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells. Tissue Eng Part A 21, 2301–2314. doi: 10.1089/ten.TEA.2014.0640 [DOI] [PubMed] [Google Scholar]
- Stein JD, Challa P, 2007. Mechanisms of action and efficacy of argon laser trabeculoplasty and selective laser trabeculoplasty. Curr Opin Ophthalmol 18, 140–145. doi: 10.1097/ICU.0b013e328086aebf [DOI] [PubMed] [Google Scholar]
- Stone EM, 2006. A very effective treatment for neovascular macular degeneration. N. Engl. J. Med. 355, 1493–1495. doi: 10.1056/NEJMe068191 [DOI] [PubMed] [Google Scholar]
- Stone EM, Andorf JL, Whitmore SS, DeLuca AP, Giacalone JC, Streb LM, Braun TA, Mullins RF, Scheetz TE, Sheffield VC, Tucker BA, 2017. Clinically Focused Molecular Investigation of 1000 Consecutive Families with Inherited Retinal Disease. Ophthalmology 124, 1314–1331. doi: 10.1016/j.ophtha.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Ming G-L, Song H, 2015. DNA damage and repair regulate neuronal gene expression. Cell Res. 25, 993–994. doi: 10.1038/cr.2015.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Iwasaki Y, Makabe K, Kamao H, Mandai M, Shiina T, Ogasawara K, Hirami Y, Kurimoto Y, Takahashi M, 2016. Successful Transplantation of Retinal Pigment Epithelial Cells from MHC Homozygote iPSCs in MHC-Matched Models. Stem Cell Reports 7, 635–648. doi: 10.1016/j.stemcr.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F, 2015. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–106. doi: 10.1038/nbt.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S, 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tan E, Wang Q, Quiambao AB, Xu X, Qtaishat NM, Peachey NS, Lem J, Fliesler SJ, Pepperberg DR, Naash MI, Al-Ubaidi MR, 2001. The relationship between opsin overexpression and photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 42, 589–600. [PubMed] [Google Scholar]
- Tang Z-H, Chen J-R, Zheng J, Shi H-S, Ding J, Qian X-D, Zhang C, Chen J-L, Wang C-C, Li L, Chen J-Z, Yin S-K, Huang T-S, Chen P, Guan M-X, Wang J-F, 2016. Genetic Correction of Induced Pluripotent Stem Cells From a Deaf Patient With MYO7A Mutation Results in Morphologic and Functional Recovery of the Derived Hair Cell-Like Cells. Stem Cells Transl Med 5, 561–571. doi: 10.5966/sctm.2015-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, Albuquerque RJC, Hauswirth WW, Chiodo VA, Kugel JF, Goodrich JA, Ponicsan SL, Chaudhuri G, Murphy MP, Dunaief JL, Ambati BK, Ogura Y, Yoo JW, Lee D-K, Provost P, Hinton DR, Núñez G, Baffi JZ, Kleinman ME, Ambati J, 2012. DICER1 Loss and Alu RNA Induce Age-Related Macular Degeneration via the NLRP3 Inflammasome and MyD88. Cell 149, 847–859. doi: 10.1016/j.cell.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa F, Maguire AM, Rossi S, Pierce EA, Melillo P, Marshall K, Banfi S, Surace EM, Sun J, Acerra C, Wright JF, Wellman J, High KA, Auricchio A, Bennett J, Simonelli F, 2013. Three-year follow-up after unilateral subretinal delivery of adeno-associated virus in patients with Leber congenital Amaurosis type 2. Ophthalmology 120, 1283–1291. doi: 10.1016/j.ophtha.2012.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK, 2017. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614. doi: 10.1038/nmeth.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Topkar VV, Joung JK, Aryee MJ, 2016. Open-source guideseq software for analysis of GUIDE-seq data. Nat. Biotechnol. 34, 483–483. doi: 10.1038/nbt.3534 [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK, 2014. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. doi: 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova B, Peng L, Liang X, Li K, Yang J-P, Ho T, Shirley J, Xu L, Potter J, Kudlicki W, Peterson T, Katzen F, 2011. Genetic assembly tools for synthetic biology. Meth. Enzymol. 498, 327–348. doi: 10.1016/B978-0-12-385120-8.00014-0 [DOI] [PubMed] [Google Scholar]
- Tucker BA, Anfinson KR, Mullins RF, Stone EM, Young MJ, 2013a. Use of a synthetic xeno-free culture substrate for induced pluripotent stem cell induction and retinal differentiation. Stem Cells Transl Med 2, 16–24. doi: 10.5966/sctm.2012-0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Mullins RF, Stone EM, 2014. Stem cells for investigation and treatment of inherited retinal disease. Hum. Mol. Genet. 23, R9–R16. doi: 10.1093/hmg/ddu124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Mullins RF, Streb LM, Anfinson K, Eyestone ME, Kaalberg E, Riker MJ, Drack AV, Braun TA, Stone EM, 2013b. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. Elife 2, e00824. doi: 10.7554/eLife.00824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Park I-H, Qi SD, Klassen HJ, Jiang C, Yao J, Redenti S, Daley GQ, Young MJ, 2011a. Transplantation of adult mouse iPS cell-derived photoreceptor precursors restores retinal structure and function in degenerative mice. PLoS ONE 6, e18992. doi: 10.1371/journal.pone.0018992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Redenti SM, Jiang C, Swift JS, Klassen HJ, Smith ME, Wnek GE, Young MJ, 2010. The use of progenitor cell/biodegradable MMP2-PLGA polymer constructs to enhance cellular integration and retinal repopulation. Biomaterials 31, 9–19. doi: 10.1016/j.biomaterials.2009.09.015 [DOI] [PubMed] [Google Scholar]
- Tucker BA, Scheetz TE, Mullins RF, DeLuca AP, Hoffmann JM, Johnston RM, Jacobson SG, Sheffield VC, Stone EM, 2011b. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 108, E569–76. doi: 10.1073/pnas.1108918108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD, 2010. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646. doi: 10.1038/nrg2842 [DOI] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K, 2014. Low Incidence of Off-Target Mutations in Individual CRISPR-Cas9 and TALEN Targeted Human Stem Cell Clones Detected by Whole-Genome Sequencing. Cell Stem Cell 15, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JA, Reynolds T, Moran ML, Moss RB, Wine JJ, 1998. Efficient and persistent gene transfer of AAV-CFTR in maxillary sinus. The Lancet. [DOI] [PubMed] [Google Scholar]
- Wang J, DeClercq JJ, Hayward SB, Li PW-L, Shivak DA, Gregory PD, Lee G, Holmes MC, 2016a. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res. 44, e30–e30. doi: 10.1093/nar/gkv1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Westenskow PD, Fang M, Friedlander M, Siuzdak G, 2016b. Quantitative metabolomics of photoreceptor degeneration and the effects of stem cell-derived retinal pigment epithelium transplantation. Philos Trans A Math Phys Eng Sci 374, 20150376. doi: 10.1098/rsta.2015.0376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin M, Pedrosa E, Hrabovsky A, Zhang Z, Guo W, Lachman HM, Zheng D, 2015. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol Autism 6, 55. doi: 10.1186/s13229-015-0048-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weleber RG, Pennesi ME, Wilson DJ, Kaushal S, Erker LR, Jensen L, McBride MT, Flotte TR, Humphries M, Calcedo R, Hauswirth WW, Chulay JD, Stout JT, 2016. Results at 2 Years after Gene Therapy for RPE65-Deficient Leber Congenital Amaurosis and Severe Early-Childhood-Onset Retinal Dystrophy. Ophthalmology 123, 1606–1620. doi: 10.1016/j.ophtha.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Whitcup SM, Sodhi A, Atkinson JP, Holers VM, Sinha D, Rohrer B, Dick A 2013. The role of the immune response in age-related macular degeneration. International journal of Inflammation. 10.1155/2013/348092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA, 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338. doi: 10.1038/nature10886 [DOI] [PubMed] [Google Scholar]
- Wilems TS, Pardieck J, Iyer N, Sakiyama-Elbert SE, 2015. Combination therapy of stem cell derived neural progenitors and drug delivery of anti-inhibitory molecules for spinal cord injury. Acta Biomater 28, 23–32. doi: 10.1016/j.actbio.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilems TS, Sakiyama-Elbert SE, 2015. Sustained dual drug delivery of anti-inhibitory molecules for treatment of spinal cord injury. J Control Release 213, 103–111. doi: 10.1016/j.jconrel.2015.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley LA, Burnight E, Kaalberg EE, Jiao C, Riker MJ, Halder JA, Luse MA, Han IC, Russell SR, Sohn EH, Stone E, Tucker BA, Mullins RF, 2017. ASSESSMENT OF AAV SEROTYPE TROPISM IN HUMAN RETINAL EXPLANTS. Hum. Gene Ther. hum.2017.179. doi: 10.1089/hum.2017.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley LA, Burnight ER, DeLuca AP, Anfinson KR, Cranston CM, Kaalberg EE, Penticoff JA, Affatigato LM, Mullins RF, Stone EM, Tucker BA, 2016. cGMP production of patient-specific iPSCs and photoreceptor precursor cells to treat retinal degenerative blindness. Sci Rep 6, 30742. doi: 10.1038/srep30742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley LA, Burnight ER, Songstad AE, Drack AV, Mullins RF, Stone EM, Tucker BA, 2015. Patient-specific induced pluripotent stem cells (iPSCs) for the study and treatment of retinal degenerative diseases. Prog Retin Eye Res 44, 15–35. doi: 10.1016/j.preteyeres.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Wilson CJ, Fennell T, Bothmer A, Maeder ML, Reyon D, Cotta-Ramusino C, Fernandez CA, Marco E, Barrera LA, Jayaram H, Albright CF, Cox GF, Church GM, Myer VE, 2017. The experimental design and data interpretation in “Unexpected mutations after CRISPR Cas9 editing in vivo” by Schaefer et al. are insufficient to support the conclusions drawn by the authors. bioRxiv 153338. doi: 10.1101/153338 [DOI] [Google Scholar]
- Wong WL, Su X, Li X, Cheung CMG, Klein R, Cheng C-Y, Wong TY, 2014. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health 2, e106–16. doi: 10.1016/S2214-109X(13)70145-1 [DOI] [PubMed] [Google Scholar]
- Worgall S, Sondhi D, Hackett NR, Kosofsky B, Kekatpure MV, Neyzi N, Dyke JP, Ballon D, Heier L, Greenwald BM, Christos P, Mazumdar M, Souweidane MM, Kaplitt MG, Crystal RG, 2008. Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum. Gene Ther. 19, 463–474. doi: 10.1089/hum.2008.022 [DOI] [PubMed] [Google Scholar]
- Worthington KS, Green BJ, Rethwisch M, Wiley LA, Tucker BA, Guymon CA, Salem AK, 2016a. Neuronal Differentiation of Induced Pluripotent Stem Cells on Surfactant Templated Chitosan Hydrogels. Biomacromolecules 17, 1684–1695. doi: 10.1021/acs.biomac.6b00098 [DOI] [PubMed] [Google Scholar]
- Worthington KS, Wiley LA, Bartlett AM, Stone EM, Mullins RF, Salem AK, Guymon CA, Tucker BA, 2014. Mechanical properties of murine and porcine ocular tissues in compression. Exp. Eye Res. 121, 194–199. doi: 10.1016/j.exer.2014.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington KS, Wiley LA, Guymon CA, Salem AK, Tucker BA, 2016b. Differentiation of Induced Pluripotent Stem Cells to Neural Retinal Precursor Cells on Porous Poly-Lactic-co-Glycolic Acid Scaffolds. J Ocul Pharmacol Ther 32, 310–316. doi: 10.1089/jop.2015.0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington KS, Wiley LA, Kaalberg EE, Collins MM, Mullins RF, Stone EM, Tucker BA, 2017. Two-photon polymerization for production of human iPSC-derived retinal cell grafts. Acta Biomater 55, 385–395. doi: 10.1016/j.actbio.2017.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang H, Colosi P, 2010. Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86. doi: 10.1038/mt.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Ren C, Liu Z, Zhang T, Zhang T, Li D, Wang L, Yan Q, Guo L, Shen J, Zhang Z, 2015. Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell. Mol. Life Sci. 72, 383–399. doi: 10.1007/s00018-014-1679-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Makiyama T, Harita T, Sasaki K, Wuriyanghai Y, Hayano M, Nishiuchi S, Kohjitani H, Hirose S, Chen J, Yokoi F, Ishikawa T, Ohno S, Chonabayashi K, Motomura H, Yoshida Y, Horie M, Makita N, Kimura T, 2017. Allele-specific ablation rescues electrophysiological abnormalities in a human iPS cell model of long-QT syndrome with a CALM2 mutation. Hum. Mol. Genet. 26, 1670–1677. doi: 10.1093/hmg/ddx073 [DOI] [PubMed] [Google Scholar]
- Yan WX, Mirzazadeh R, Garnerone S, Scott D, Schneider MW, Kallas T, Custodio J, Wernersson E, Li Y, Gao L, Federova Y, Zetsche B, Zhang F, Bienko M, Crosetto N, 2017. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun 8, 15058. doi: 10.1038/ncomms15058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Grishin D, Wang G, Aach J, Zhang C-Z, Chari R, Homsy J, Cai X, Zhao Y, Fan J-B, Seidman C, Seidman J, Pu W, Church G, 2014. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun 5, 5507. doi: 10.1038/ncomms6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Güell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, Huang P-Y, Daley G, Church G, 2013. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 41, 9049–9061. doi: 10.1093/nar/gkt555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, Wilson JM, 2016. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 34, 334–338. doi: 10.1038/nbt.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Tucker BA, Zhang X, Checa-Casalengua P, Herrero-Vanrell R, Young MJ, 2011. Robust cell integration from co-transplantation of biodegradable MMP2-PLGA microspheres with retinal progenitor cells. Biomaterials 32, 1041–1050. doi: 10.1016/j.biomaterials.2010.09.063 [DOI] [PubMed] [Google Scholar]
- Yiu G, He Z, 2006. Glial inhibition of CNS axon regeneration. Nat. Rev. Neurosci. 7, 617–627. doi: 10.1038/nrn1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, Tieu E, Nguyen AT, Wong B, Smit-McBride Z, 2016. Genomic Disruption of VEGF-A Expression in Human Retinal Pigment Epithelial Cells Using CRISPR-Cas9 Endonuclease. Invest. Ophthalmol. Vis. Sci. 57, 5490–5497. doi: 10.1167/iovs.16-20296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA, 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. doi: 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim J-W, Brooks M, Ataeijannati Y, Sun X, Dong L, Li T, Swaroop A, Wu Z, 2017. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun 8, 14716. doi: 10.1038/ncomms14716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumlu S, Stumm J, Bashir S, Dreyer A-K, Lisowski P, Danner E, Kühn R, 2017. Gene editing and clonal isolation of human induced pluripotent stem cells using CRISPR/Cas9. Methods 121–122, 29–44. doi: 10.1016/j.ymeth.2017.05.009 [DOI] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F, 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-P, Li X-L, Li G-H, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu Y-W, Xu J, Chun N, Yuan W, Cheng T, Zhang X-B, 2017. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 18, 35. doi: 10.1186/s13059-017-1164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Rho M, Tang H, Doak TG, Ye Y, 2013. CRISPR-Cas systems target a diverse collection of invasive mobile genetic elements in human microbiomes. Genome Biol. 14, R40. doi: 10.1186/gb-2013-14-4-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Klassen HJ, Tucker BA, Perez M-TR, Young MJ, 2007. CNS progenitor cells promote a permissive environment for neurite outgrowth via a matrix metalloproteinase-2-dependent mechanism. J. Neurosci. 27, 4499–4506. doi: 10.1523/JNEUROSCI.0200-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhang Z-N, Westenskow PD, Todorova D, Hu Z, Lin T, Rong Z, Kim J, He J, Wang M, Clegg DO, Yang Y-G, Zhang K, Friedlander M, Xu Y, 2015. Humanized Mice Reveal Differential Immunogenicity of Cells Derived from Autologous Induced Pluripotent Stem Cells. Cell Stem Cell 17, 353–359. doi: 10.1016/j.stem.2015.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen S, Takahashi Y, Narita S, Yang Y-C, Li X, 2017. Targeted delivery of CRISPR/Cas9 to prostate cancer by modified gRNA using a flexible aptamer-cationic liposome. Oncotarget 8, 9375–9387. doi: 10.18632/oncotarget.14072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Lawrence M, Gupta A, Pagès H, Kucukural A, Garber M, Wolfe SA, 2017. GUIDEseq: a bioconductor package to analyze GUIDE-Seq datasets for CRISPR-Cas nucleases. BMC Genomics 18, 379. doi: 10.1186/s12864-017-3746-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Mali P, Huang X, Dowey SN, Cheng L, 2011. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood 118, 4599–4608. doi: 10.1182/blood-2011-02-335554 [DOI] [PMC free article] [PubMed] [Google Scholar]


