FIGURE 6.
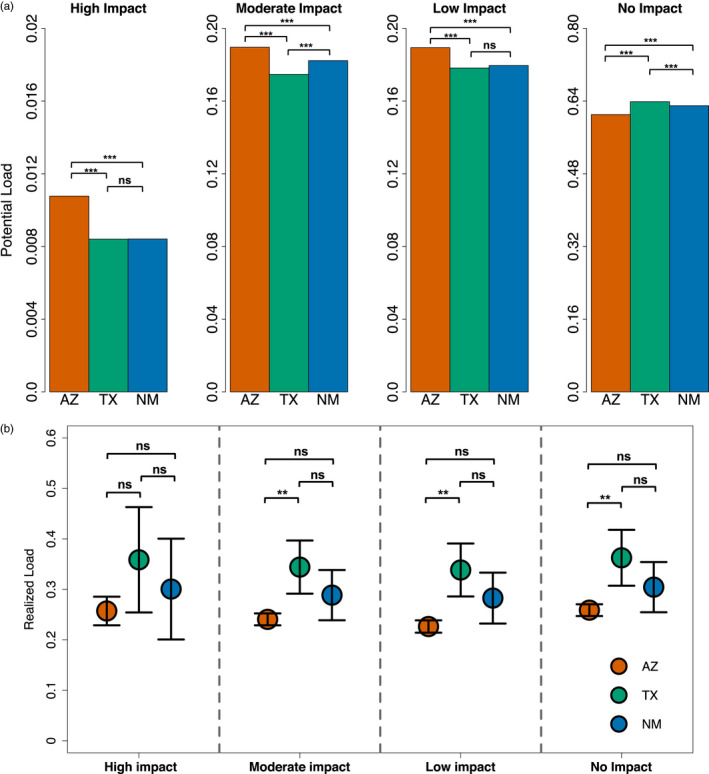
Larger populations have higher potential genetic load, but load is more realized in smaller, inbred populations (see Materials and Methods for details). Variants were classified as either high, moderate, low, or no impact based on their inferred effect on protein translation. High‐impact variants should have the most disruptive (i.e., deleterious) effect, whereas low‐ or no‐impact mutations were mostly synonymous substitutions with little to no impact on protein sequences (a) Potential genetic load was estimated for each population as the proportion of deleterious mutations within annotated protein‐coding genes. The Arizona samples had the highest potential load of high‐impact, moderate‐impact, and low‐impact variants. Note the difference in scales on y‐axis. (b) Realized load was measured as the mean frequency of deleterious alleles found within individual genomes for each impact class (N = 21; AZ = 7, TX = 7, and NM = 7). No significant difference was found in realized load of highly deleterious mutations between Texas and Arizona quail, but the small Texas population has a higher frequency of moderate‐, low‐, and no‐impact variants coupled with more inbreeding and more homozygosity (Figures 2, 3, and S9) than the larger outbred Arizona populations. Error bars indicate 95% CI around the estimates. ns p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001
