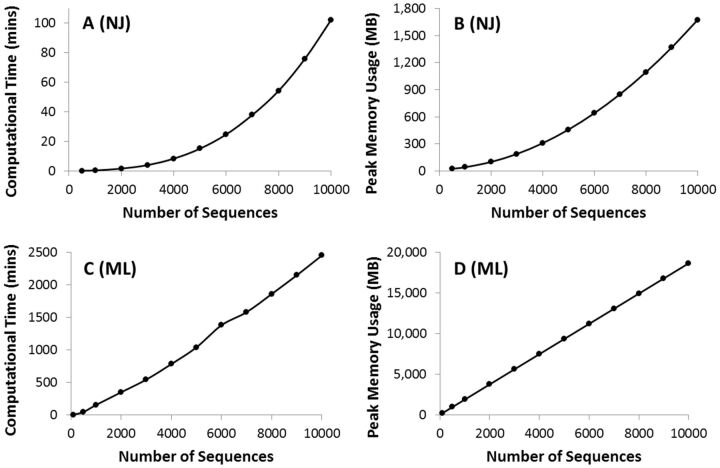
F ig . 1.
Time and memory requirements for phylogenetic analyses using the NJ method ( A , B ) and the ML analysis ( C , D ). For NJ analysis, we used the Tamura–Nei (1993) model, uniform rates of evolution among sites, and pairwise deletion option to deal with the missing data. Time usage increases polynomially with the number of sequences (third degree polynomial, R 2 = 1), as does the peak memory used ( R 2 = 1) ( A , B ). The same model and parameters were used for ML tree inference, where the time taken and the memory needs increased linearly with the number of sequences. For ML analysis, the SPR (Subtree–Pruning–Regrafting) heuristic was used for tree searching and all 5,287 sites in the sequence alignment were included. All the analyses were performed on a Dell Optiplex 9010 computer with an Intel Core-i7-3770 3.4 GHz processor, 20 GB of RAM, NVidia GeForce GT 640 graphics card, and a 64-bit Windows 7 Enterprise operating system.