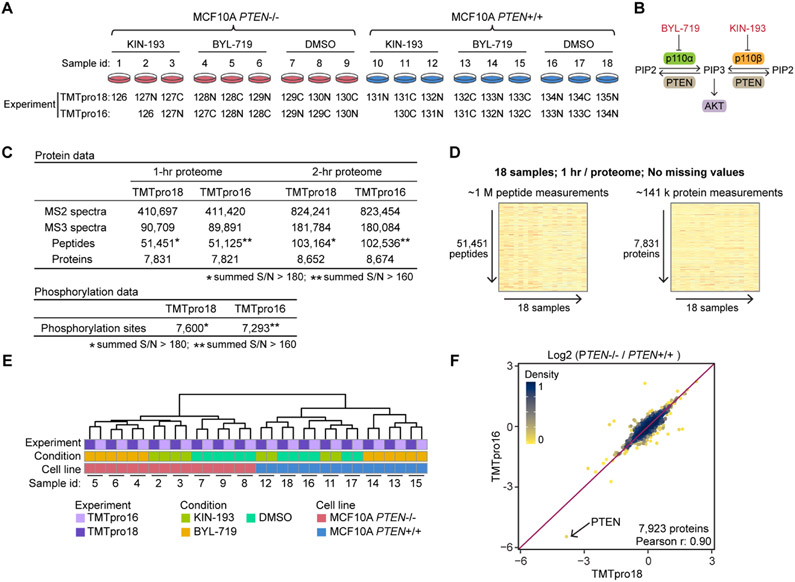
Figure 2.
TMTpro-18plex reagents facilitate proteome analysis at an effective rate of 1 h per proteome. (A) Set of 18 samples of drug-treated control MCF10A and MCF10A PTEN−/− cell lines was used to benchmark the TMTpro-18plex reagents. The cell lines were treated with a PI3K inhibitor (BYL-719 or KIN-193) or DMSO, in biological triplicate (n = 3). Samples were prepared following the SL-TMT protocol using 50 μg protein and labeling with either the full 18 reagents or only 16 reagents as indicated. Peptides were fractionated and then concatenated into 24 “super” fractions. Fractionated samples were analyzed with FAIMS and real-time search-synchronous-precursor-selection-MS3 on an Orbitrap Eclipse mass spectrometer. An equal amount of fractionated peptide was analyzed in both experiments. (B) PI3K overview. KIN-193 and BYL-719 inhibit the kinases p110α and p110β, respectively. p110α and p110β are catalytic subunits of PI3K. PI3K-mediated PIP3 production leads to the activation of the downstream AKT/mTOR pathway. (C) Data set overview. Analysis of 12 fractions (1 h per proteome) or 24 fractions (2 h per proteome) yielded ~7.8k and ~8.6k quantified proteins in both experiments, respectively. A total amount of 900 and 800 μg of peptide was used to enrich phosphorylated peptides in the 18- and 16-plex experiments, respectively. The numbers of localized and quantified phosphorylation sites are reported (AScore > 13). (D) 1 h human cell line proteome. The TMTpro-18plex reagents facilitated the collection of a 1 h per proteome analysis that included ~1 million peptide measurements and ~141,000 protein measurements across 18 samples without missing values. (E) HCA of the protein data; (F) log 2 protein fold changes between the two cell lines treated with DMSO in both experiments showed good agreement.