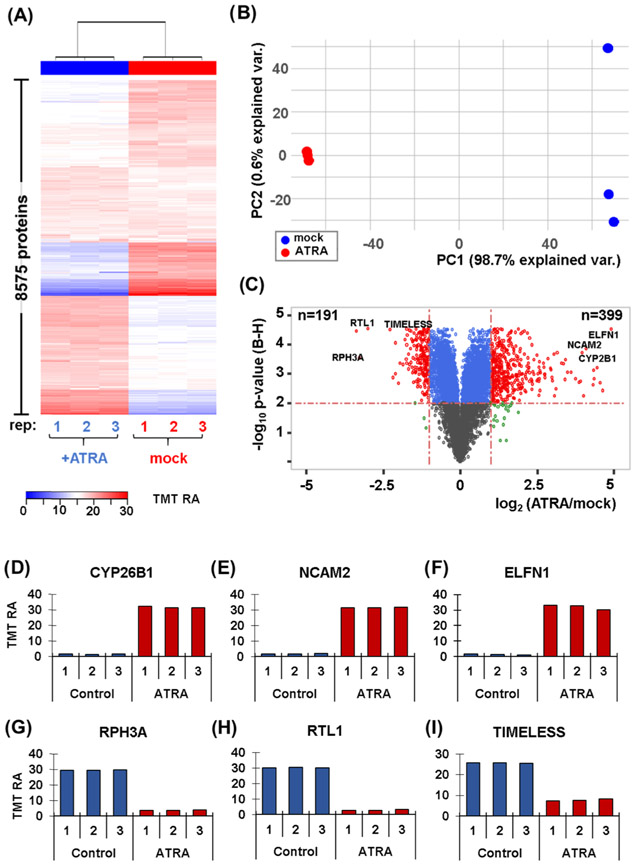
Figure 2.
Proteome profiling of SH-SY5Y cells with and without a 7-day retinoic acid treatment. (A) Heat map illustrates the relative abundance of the 8,575 proteins quantified in this experiment. Each protein abundance value sums to 100 across the six channels. (B) Principal components analysis (PCA) of the six samples depicts the variance explained by the first two principal components (PC). (C) Volcano plot illustrating the Benjamini–Hochberg (BH)-corrected p-value of retinoic acid (ATRA) vs. mock control and the fold change for these values. Highlighted on the volcano plot are six of the most altered proteins resulting from retinoic acid treatment. These proteins include: (D) CYP26B1, cytochrome P450 26B1; (E) NCAM2, neural cell adhesion molecule 2; (F) ELFN1, extracellular leucine-rich repeat and fibronectin type III.(G) RPH3A, rabphilin-3A; (H) RTL1, retrotransposon-like protein 1; and (I) TIMELESS.