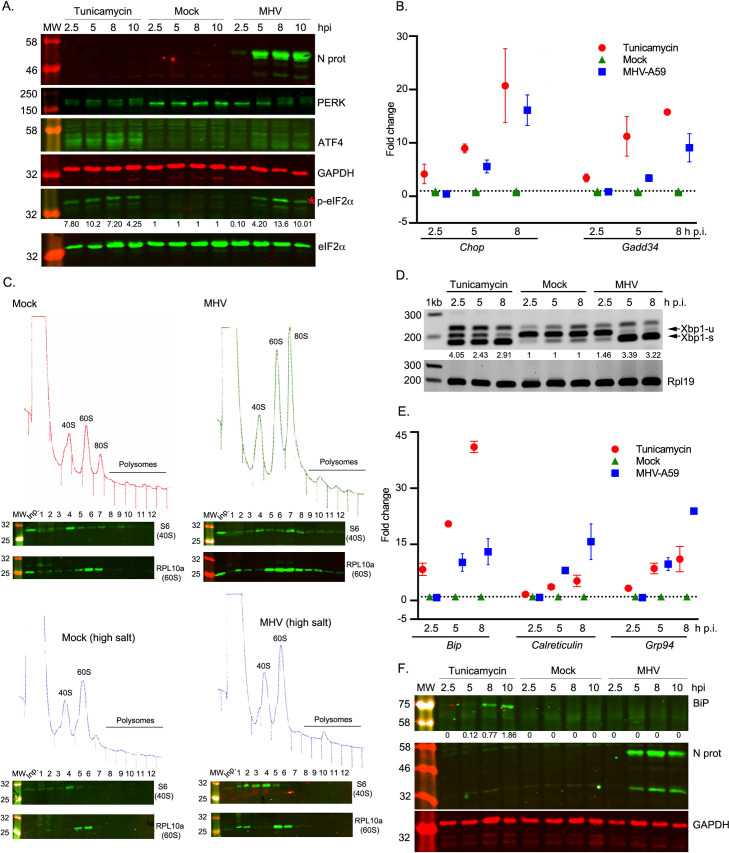
Fig 2. MHV infection and activation of the unfolded protein response.
17 Cl-1 cells were incubated in the presence of tunicamycin (2 μg/ml) or infected with MHV-A59 (MOI 5) and harvested at 2.5, 5, 8 and 10 h p.i. (A) Western blot analysis of ATF4, p-eIF2α, eIF2α, PERK and MHV N proteins. GAPDH and eIF2α were used as loading controls. Molecular masses (kDa) are indicated on the left and the p-eIF2α band is indicated by a red asterisk. Protein band quantifications for p-eIF2α, normalised by eIF2α and given relative to the timepoint-matched mock value, are provided below the immunoblot. (B) RT-qPCR of Chop and Gadd34 mRNA for three biological replicates of a timecourse of MHV infection or tunicamycin treatment. Data are normalised using Rpl19 as a housekeeping gene and presented as fold change of expression relative to mock-infected cells (marked as a dashed line). (C) Mock-infected (left upper panel) and MHV-infected (right upper panel) 17 Cl-1 cells were harvested at 5 h p.i. Cytoplasmic lysates were resolved on 10–50% sucrose density gradients. Gradients were fractionated and fractions monitored by absorbance (A254 nm). Twelve numbered fractions were collected and proteins extracted, resolved by 12% SDS-PAGE and analysed by immunoblotting using the indicated antibodies (anti-S6 as 40S marker, anti-RPL10 as 60S marker). Mock-infected (left lower panel) and MHV-infected (right lower panel) 17 Cl-1 cells were harvested at 5 h p.i. in high-salt lysis buffer (400 mM KCl) and analysed as described above. Lane "Inp" contains whole cell lysate. (D) RT-PCR analysis of Xbp1-u and Xbp1-s mRNAs. Rpl19 RT-PCR product was used as a loading control. Molecular size markers (nt) are indicated on the left. Xbp1 splicing was quantified as the ratio Xbp1-s / (Xbp1-s + Xbp1-u), and the extent of splicing relative to the timepoint-matched mock is shown below each lane. The band that migrates above Xbp1-u is thought to represent a duplex of Xbp1-s and Xbp1-u, known as the “hybrid” band (Xbp1-h) [116]. (E) RT-qPCR of Bip, Calreticulin and Grp94 mRNA for three biological replicates of a timecourse of MHV infection or tunicamycin treatment. Data are normalised as in B. (F) Cell lysates were analysed by 12% SDS-PAGE and immunoblotted using anti-BiP and anti-N antibodies. GAPDH was used as a loading control. Protein band quantifications for BiP were normalised to GAPDH. Immunoblots and agarose gels are representative of three biological replicates.