Fig. 1.
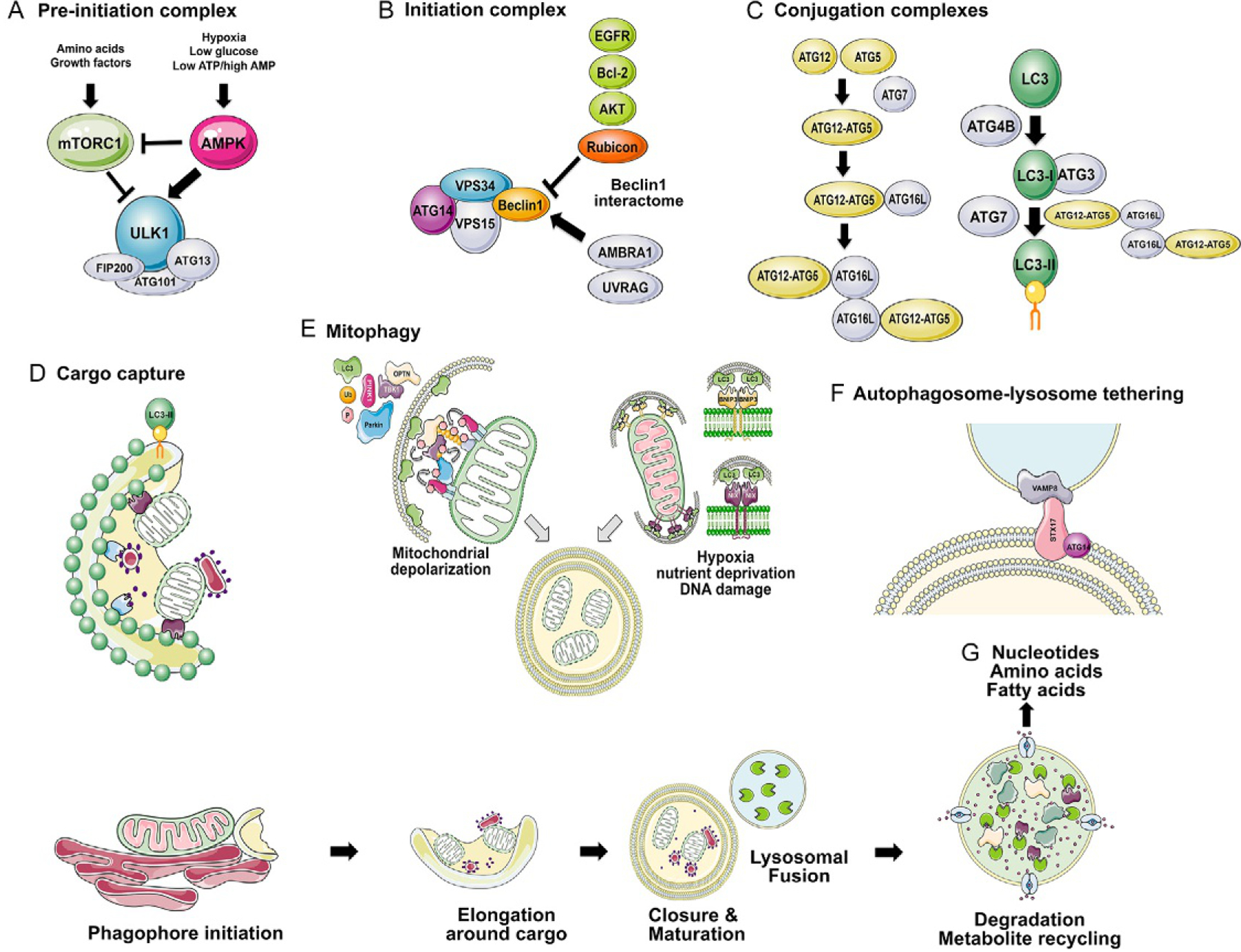
Overview of the molecular basis of autophagy initiation and completion. The process of autophagy is summarized above with (A) nutrient control of the preinitiation complex (containing the critical ULK1/ULK2, FIP200, ATG13 and ATG101) via activation of AMPK and repression of mTOR; (B) initiation complex made up of VPS34, VPS15, ATG14 and Beclin1 promotes phagophore formation and recruitment of the conjugation complexes, ATG12-ATG5/ATG16L and LC3-II-PE; (C) conjugation complexes ATG12-ATG5/ATG16L and LC3-II-PE are formed under the control of ATG7; (D) autophagic cargo is recruited to the growing phagophore through interaction of cognate cargo receptors, such as NIX (purple) with mitochondria and NUPIF1 (pale blue) with ribosomes via LIR motifs with LC3-II; (E) mitophagy is a selective form of autophagy in which mitochondria are targeted for degradation as a result of depolarization in a ubiquitin-dependent manner (PINK1/Parkin), or in response to nutrient stress in a ubiquitin-independent manner (BNIP3/NIX); (F) fusion of the autophagosome with the lysosome is promoted by the interaction of STX17 together with ATG14 at the autophagosome, with Vamp8 at the lysosome; (G) autophagic cargo is degraded at the lysosome via lysosomal hydrolases.
