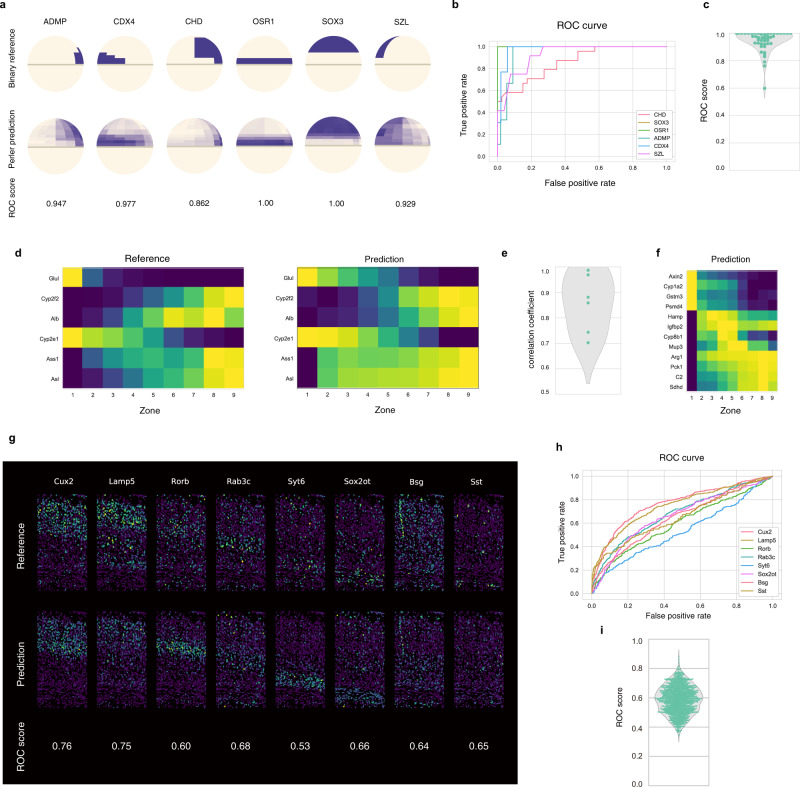
Fig. 6. Applications of Perler to other data.
a–c Application of Perler for early zebrafish embryo data. a LOOCV experiments. The upper and lower panels show the referenced ISH data and predicted gene-expression profiles, respectively. b ROC curve for the LOOCV experiments for the genes shown in a. c Violin plot for the predictive accuracies of Perler for the LOOCV experiments for all genes in the reference ISH data according to ROC score. d–f Application of Perler for mammalian liver lobules. d LOOCV experiments. The left and right panels show the reference ISH data and predicted gene-expression profiles, respectively. All genes from the ISH data are shown. e Violin plot for the predictive accuracies of Perler for the LOOCV experiments for all genes in the reference ISH data. f Prediction of non-landmark genes. In addition to the monotonic gene-expression profiles, non-monotonic gene-expression profiles are observed (Hamp, Igfbp2, Cyp8b1, and Mup3)7. g, h Application of Perler for the mouse visual cortex. g Cross-validation (CV) study (tenfold). The upper and lower panels show the referenced ISH data and predicted gene-expression profiles, respectively. h ROC curve for the CV experiments of genes shown in g. i Violin plot representing the predictive accuracies of Perler for the tenfold CV experiments of all genes in the reference ISH dataset according to ROC score. The median ROC score is 0.59. Source data are provided in a Source data file.