Figure 1 .
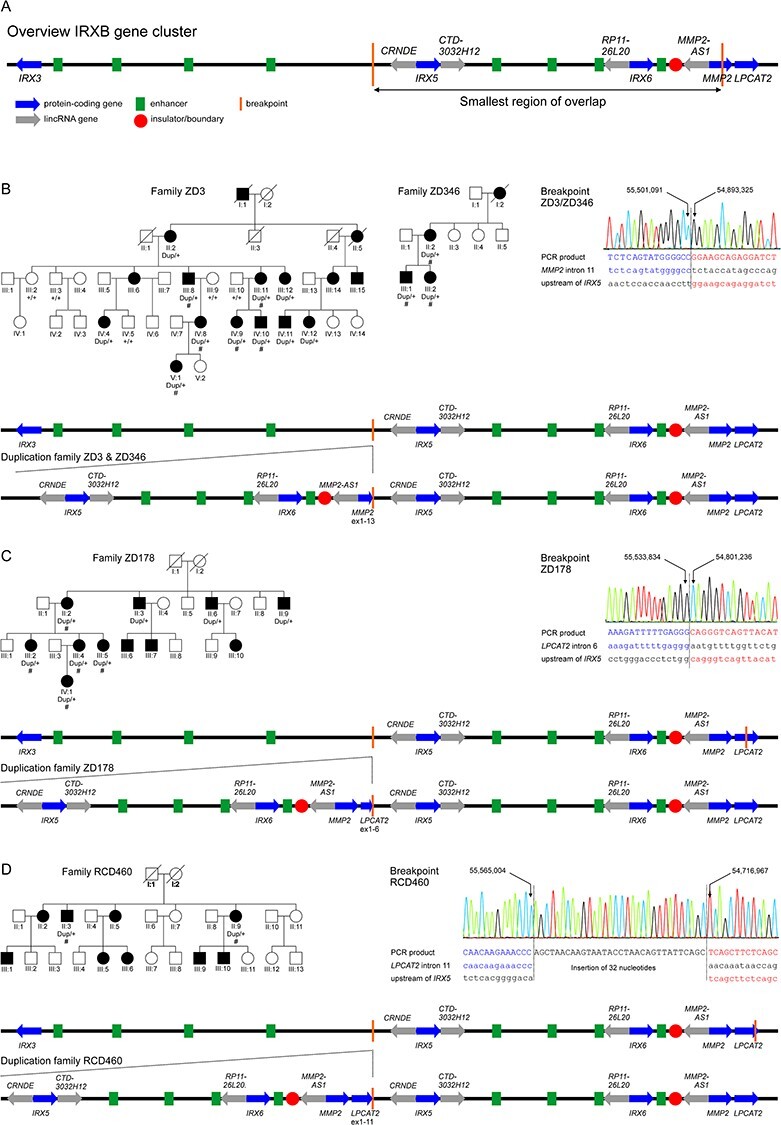
Tandem duplications at the IRXB gene cluster on chromosome 16q12 segregate with autosomal dominant cone dystrophy in four families. (A) Genomic organization of the IRXB gene cluster and the other two genes involved in the duplications, MMP2 and LPCAT2. Blue arrows indicate the protein-coding genes IRX3, IRX5, IRX6, MMP2 and LPCAT2, and gray arrows the non-coding RNAs CRNDE, CTD-3032H12, RP11-26 L20 and MMP2-AS1. The IRXB locus is highly conserved throughout evolution, not only in its genes but also its regulatory elements. Conserved enhancer elements are depicted in green and insulators/boundaries are represented with a red circle (4). The smallest region of overlap of the duplications found in this study is depicted as by vertical orange lines. (B–D) Pedigrees and segregating genotypes for the analyzed subjects and families in this study, breakpoint mapping and genomic organization of the duplication. Pedigrees: Patients for which DNA was available for CNV analysis are indicated by the identified genotype beneath the individual identifier. The presence of the duplication is indicated by ``dup'', while ``+'' indicates normal wild-type alleles. Patients for which clinical data was available are presented in Figure 3, Table 2 and Supplementary Material, Table S1 and are indicated by ``#''. Circles depict females and squares males, affected individuals by filled and unaffected individuals by open symbols. Already deceased individuals are marked by a diagonal slash. Breakpoint mapping in the investigated families: Electropherograms of the breakpoint sequences as obtained from Sanger sequencing of PCR products covering the breakpoints. Left and right junction sequences are given in blue and red, respectively. The reference sequences are given in lowercase letters below the sequence for the PCR product. Genomic positions refer to GRCh38/hg38. Schematic representation of the tandem repeated duplication as observed in the families in this study in comparison to the genomic organization of the wild-type IRXB gene cluster in families ZD3 and ZD346 (B), family ZD178 (C) and family RCD460 (D). The tandem repeated sequence is expected to create a new topological associated domain (TAD) flanked by the insulators as boundaries.
