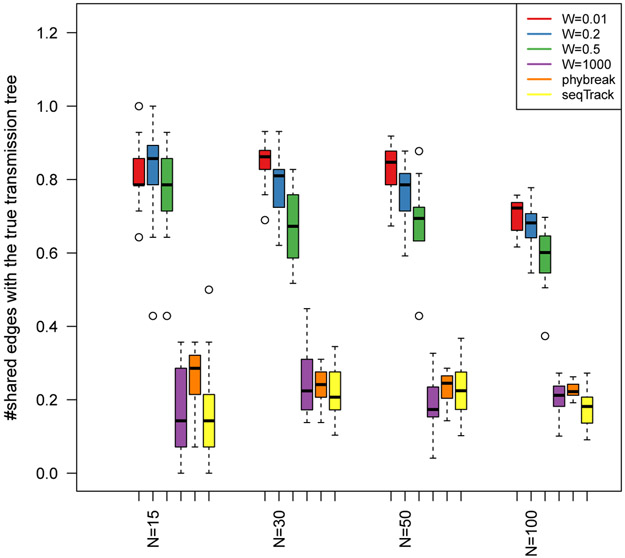
Fig 3.
Performance of the proposed MCMC sampler for different sizes of transmission trees (N) and various infection time intervals (specified by W). Performance is also compared to two alternative reconstruction methods phybreak and seqTrack, which do not take into account infection intervals. The proposed method is able to incorporate this additional information in the estimation. According to this figure, availability of more accurate data on infection times leads to more accurate reconstruction of transmission trees. The proposed method with large W, the phybreak and seqTrack methods provide highly unreliable estimates.