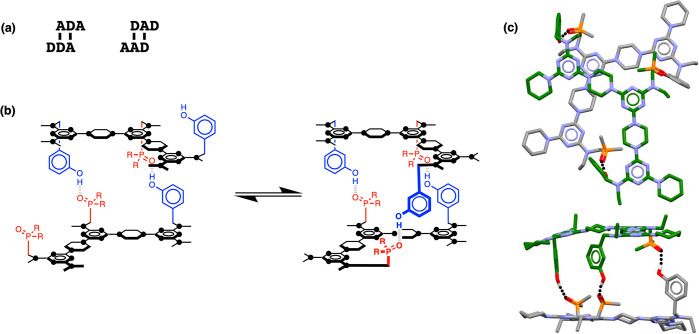
Figure 11.
(a) Two mismatched duplexes that are expected to make two H-bonds when the sequences are arranged in a linear fashion but actually form three H-bonds based on ITC measurements. (b) Structure of the DAD·AAD duplex showing how the terminal dangling bases (left) can fold back to make a third H-bond (right). The melamine nitrogen atoms are represented as black dots, and the terminal piperidine and solubilizing groups are not shown. (c) Top and side views of the energy minimum structure of the DAD·AAD duplex (MMFFs with chloroform solvation). H-bonds are indicated by black dotted lines. The solubilizing groups and hydrogen atoms are omitted for clarity