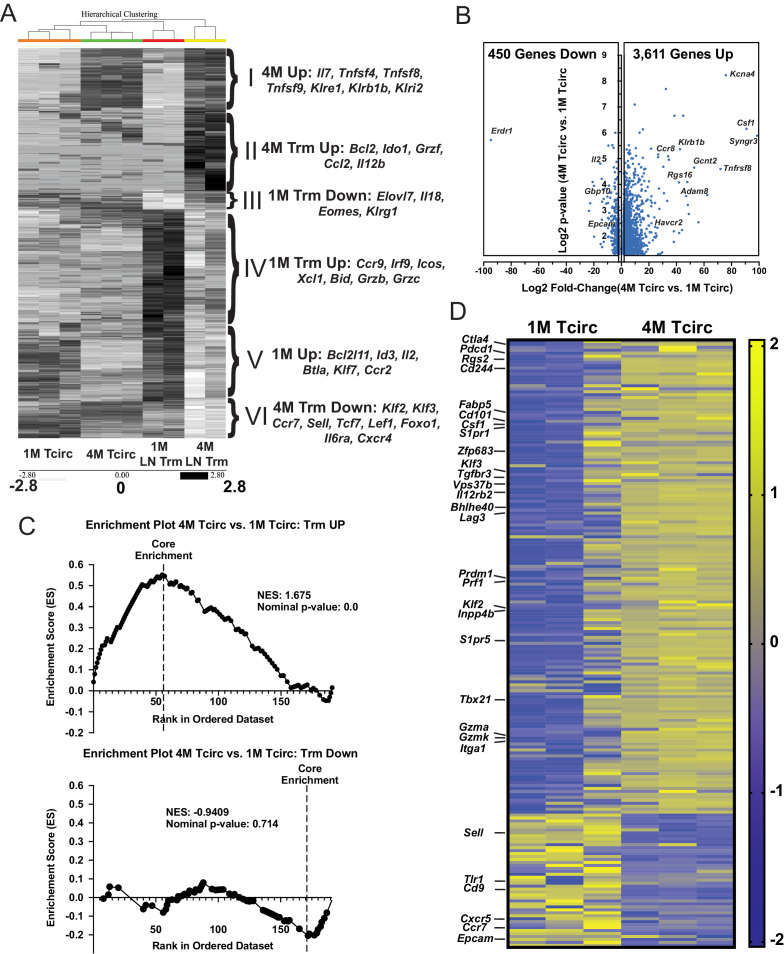
Figure 7. Splenic 4M cells express a core Trm signature.
Mice were seeded with 104 naive P14 or 105 3M P14 cells and IN infected with PR8-GP33 virus. At 22–30 days post-infection, IV exclusion was performed and negatively enriched pooled groups of spleens (3–5 spleens/sample, n = 3) or mLNs (15–25 mLNs/sample, n = 2) were stained for CD8α, CD90.1, CD69, and CD103. Bulk RNAseq was performed on RNA from sort-purified spleen samples (20k IV−, CD69−/CD103− cells/sample) or mLN Trm samples (2–5k IV− CD69+/CD103+ cells/sample). (A) Heatmaps of 1300 most differentially expressed genes (log2FC > 1.5, p<0.05) between 1M and 4M LN Trms are plotted from the four respective groups of samples. The six core signature sets of genes offset to the right were derived from unbiased hierarchical clustering. (B) Volcano plot of 4061 differentially expressed genes between 1M and 4M splenic memory P14 cells (log2FC > 1.5, p<0.05). (C) GSEA of core Trm genes defined in Table 2 from splenic 1M and 4M populations separated into respective upregulated (top) and downregulated (bottom) gene sets in regard to annotated expression in Trms. (D) Heatmap of a core set of selected Trm genes (as in C) within 1M and 4M splenic populations.