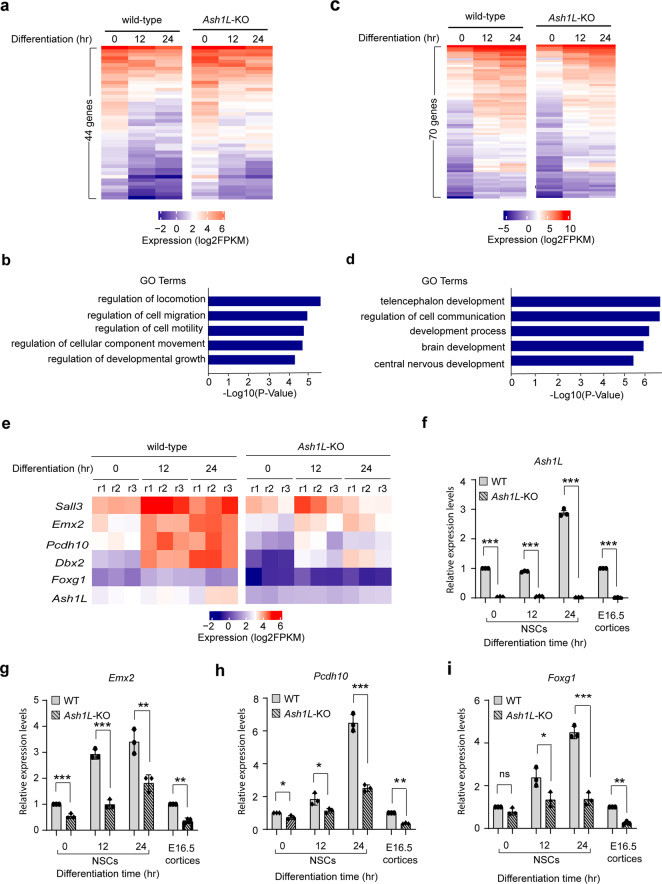
Fig. 4. Loss of ASH1L impairs expression of genes critical for brain development.
a Heatmap showing 44 genes downregulated in the differentiating wild-type NPCs have significant increased expression in the Ash1L-KO cells. b Gene ontology enrichment analysis showing the enriched GO terms of 44 genes upregulated in the Ash1L-KO cells during differentiation (FDR < 0.05). c Heatmap showing 70 genes upregulated in the differentiating wild-type NPCs have significant reduced expression in the Ash1L-KO cells. d Gene ontology enrichment analysis showing the enriched GO terms of 70 genes downregulated in the Ash1L-KO cells during differentiation (FDR < 0.05). e Heatmap showing the representative NDD-related genes downregulated in the Ash1L-KO cells during differentiation (cutoff: fold change > 1.5, p < 0.01). f–i qRT-PCR analysis showing the mRNA levels of Ash1L, Emx2, Pcdh10, and Foxg1 in wild-type and Ash1L-KO NPCs at different time points of induced differentiation and in bulk E16.5 cortices. The results of analysis in NPCs were normalized against levels of Gapdh and the expression level of wild-type NPCs at differentiation time 0 was arbitrarily set to 1. The results of analysis in E16.5 cortices were normalized against levels of Gapdh and the expression level of wild-type cortices was arbitrarily set to 1. P-values calculated using a two-tailed t test. The error bars represent mean ± SEM, n = 3 biologically independent samples/genotype. Note: *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.