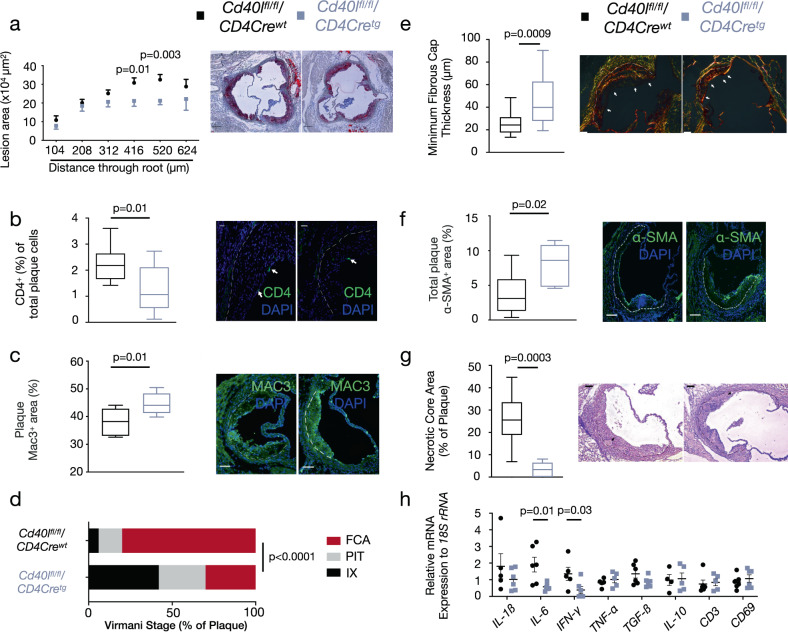
Fig. 1. Lack of T cell CD40L reduces plaque burden with less advanced, more stable plaques.
a Atherosclerotic plaque area in cross-sections at indicated positions of the aortic root from Cd40lfl/fl/Cd4Cre mice [n: WT = 17, TG = 16] with representative Oil Red O-stained photomicrographs (scale bar: 200 µm). b, c Immunofluorescent staining assessing plaque phenotypes in cross-sections of the aortic root of mice with quantifications (left) and representative photomicrographs (right). Arrows indicate stained cells and the dashed line separates the atherosclerotic lesion. b CD4+ T cells (green) along with nuclear DAPI staining (blue) [n = 10] and (c) Mac3+ area (green) along with nuclear DAPI staining (blue) [n = 7] (scale bar: 100 µm). d Chi-square distribution analysis of morphological plaque phenotypes describing initial xanthoma (IX), pathological intimal thickening (PIT), and fibrous cap atheroma (FCA) phenotypes [n: WT = 17 animals/51 lesions, TG = 13/39 lesions]. e–g Multi-parameter analysis of plaque stability assessing: e Fibrous cap thickness [n: WT = 20, TG = 14] using a Sirius Red stain (polarized light), (f) Alpha-smooth muscle actin+ (α-SMA+) area [n: WT = 9, TG = 5] (green, scale bar: 75 µm), and (g) Necrotic core content [n = 8] using hematoxylin and Eosin stain all in aortic root sections with quantifications (left) and representative photomicrographs (right). h Gene expression analysis using qPCR to evaluate various mRNA cytokine expression profiles in the descending aorta [n: WT = 6: CD3, TGF-ß, IL-6, CD69; 5: IL-1ß, 4: IFN- γ, IL-10, TNF- α; TG = 7: TGF-ß; 6: IL-6, CD69; 5: IL-1ß, IL-10, IFN- γ, CD3; 4: TNF- α. Data are represented as either mean ± s.e.m. a, h or boxplots (b, c, e–g). In the boxplots, whiskers above and below the boxes indicate the 10th and 90th percentile while the boundaries of the boxes represent the 25th and 75th percentile. The black line within the boxes marks the median. n refers to biologically independent animals. Data were analyzed by either a two-tailed unpaired Student’s t test (a–c, f), two-tailed Mann–Whitney test (e, g, h), or Chi-square test (d). Interleukin-1 beta IL-1ß, Interleukin-6 IL-6, Interferon-gamma IFN-γ, Tumor necrosis factor-alpha TNF-α, Transforming growth factor-beta TGB-ß, Interleukin-10 IL-10, Cluster of differentiation 3 CD3. Source data are provided as a Source Data file.