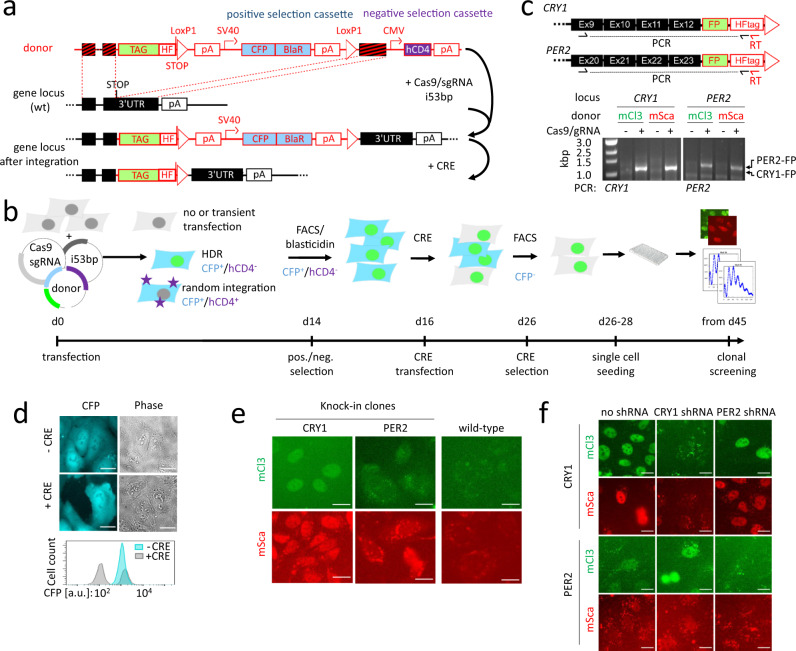
Fig. 1. CRISPR/Cas9-mediated generation of clock protein knock-in reporter cells.
a Donor plasmid design and genome editing strategy: The tag (e.g. fluorescent protein) to be integrated and a floxed positive selection cassette (cyan fluorescent protein (CFP) + blasticidin-resistance (BlaR)) are flanked by arms, which are homologous to the genomic target region. When Cas9/single guide RNA(sgRNA)-mediated DNA double strand breaks are repaired by HDR, tag, and positive selection marker are integrated into the target region. The negative selection cassette (hCD4, a cell surface protein exclusively expressed on immune cells) is only integrated into the genome by unwanted random integration of the whole donor plasmid. b Selection strategy. Cells are transfected with Cas9, sgRNA, i53bp (see text), and donor plasmid. Stable transfectants are selected by blasticidin selection and fluorescence-activated cell sorting (FACS) of CFP positive cells (blue), while unwanted hCD4 positive cells (purple stars) are depleted. Subsequently, cells are transiently transfected with Cre recombinase (CRE) to remove the positive selection cassette from the genomic locus, and only CFP negative cells (gray) are clonally expanded and screened for successful knock-in. c Chimeric mRNA was detected in selected batch cultures by reverse transcription-polymerase chain reaction (RT-PCR) using a RT- and a reverse primer specific to the insertion and a gene-specific forward primer. d Loss of CFP expression after removal of the positive selection cassette monitored by microscopy and flow cytometry. The gating strategy used FSC and SSC signals to gate out doublets and debris. e Fluorescence microscopy images of successful knock-in clones. f Indicated knock-in cells were either left untreated or transduced with shRNA targeting either CRY1 or PER2. Images were acquired 10 h after synchronization. Corresponding differential interference contrast (DIC) images are shown in Supplementary Fig. 3a. Scale bars: 20 µm. mCl3 mClover3, mSca mScarlet-I, FP fluorescent protein, pA polyadenylation signal, SV40 simian virus-40 promoter, CMV cytomegalovirus promoter, HF His-tag/FLAG-tag, shRNA short hairpin RNA, UTR untranslated region, Ex exon, FSC forward scatter, SSC sideward scatter. Source data are provided as a Source Data file.