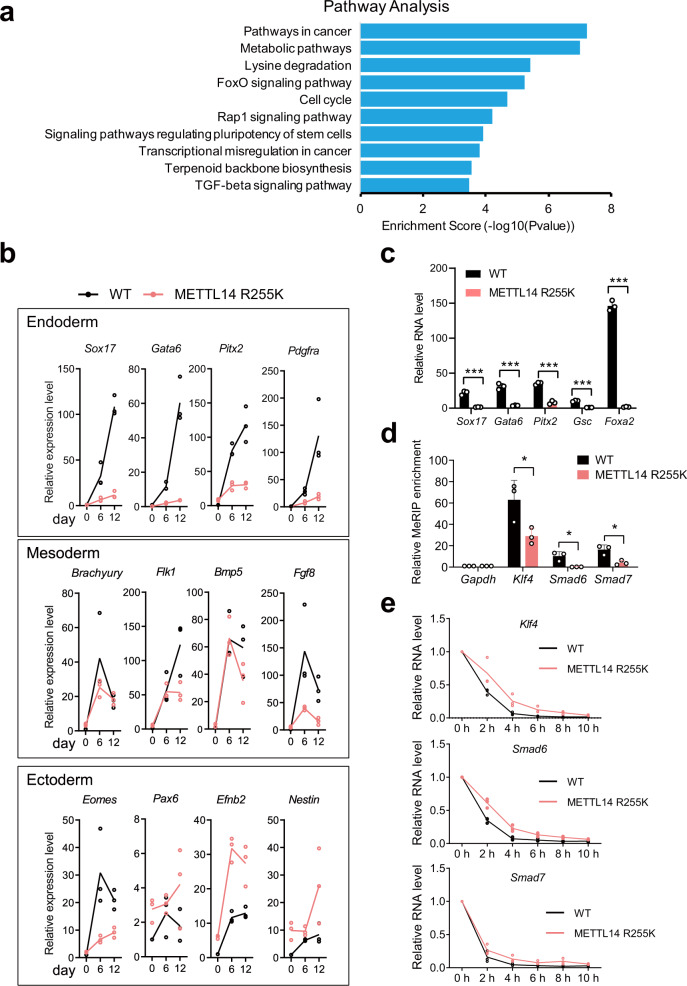
Fig. 6. Methylation of METTL14 at arginine 255 is required for normal mESC endoderm differentiation.
a KEGG pathway enrichment analysis of genes associated with downregulated m6A regions in R255K cells. One-sided hypergeometric test, P value was adjusted by Benjamini and Hochberg method. b Quantitative RT-PCR analysis of differentiation markers of ectoderm, mesoderm, and endoderm in WT and R255K mESCs. The expression level was normalized against Gapdh then to WT at day 0. Data from three independent experiments are shown. c Quantitative RT-PCR analysis of markers after directed differentiation to definitive endoderm fate using FGF2 and activin A in WT or R255K mESCs. The expression of genes was first normalized to Gapdh then to that in undifferentiated mESCs. Data are mean ± s.d. from three independent experiments. Two-sided Student’s t-test. d MeRIP-qPCR of m6A peaks on Klf4, Smad6, and Smad7. Enrichment of MeRIP versus input RNA was normalized against Gapdh. Data are mean ± s.d. from three independent experiments. Two-sided Student’s t-test. e The stability of Klf4, Smad6, and Smad7 RNAs in WT and R255K cells after Actinomycin D treatment. The expression level was normalized against Gapdh then to WT at 0 h. Data from three independent experiments are shown. Source data for (b–e) are provided as a Source Data file.