Fig 1.
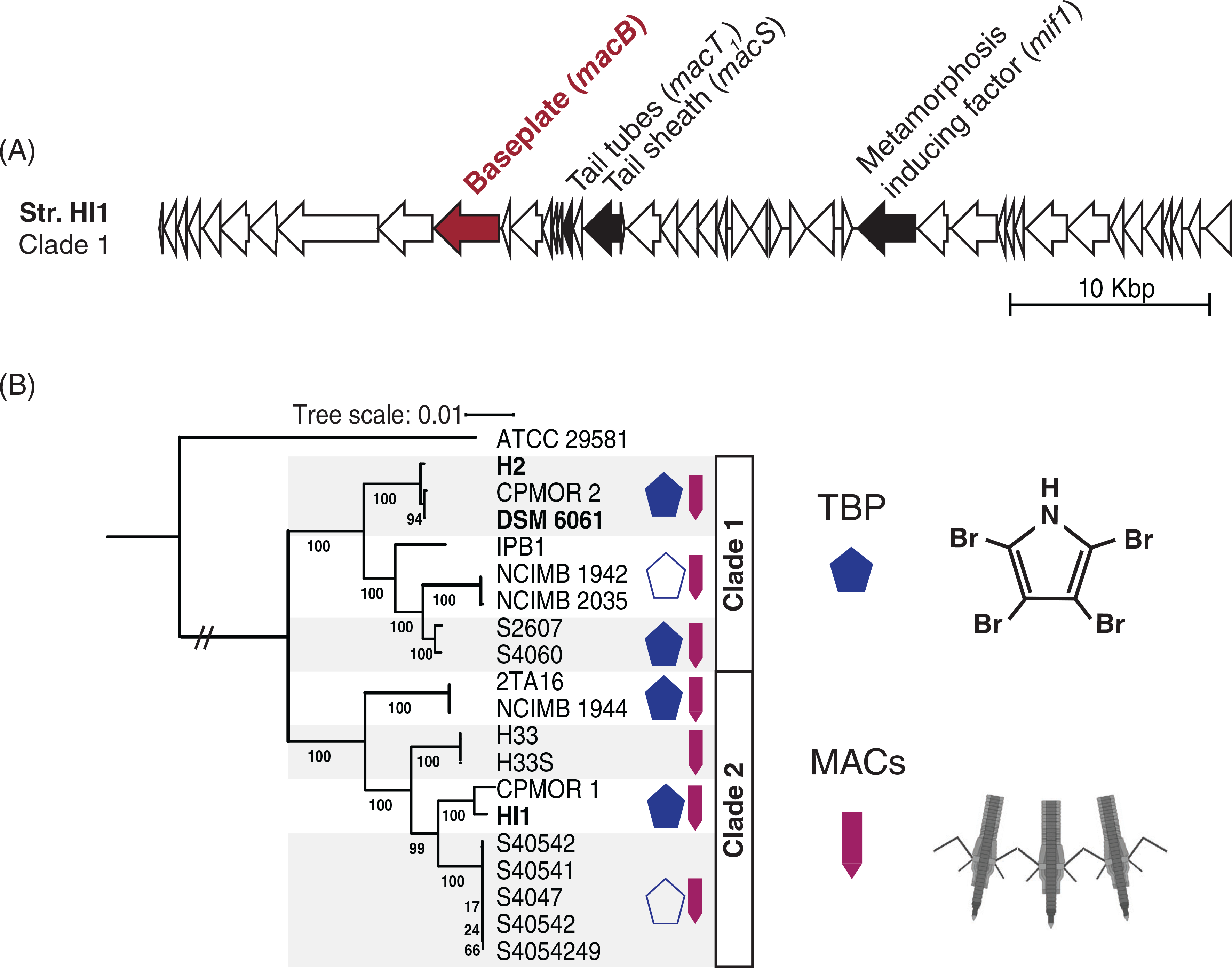
Diverse Pseudoalteromonas luteoviolacea strains encode the biosynthesis genes for TBP and MACs. (A) MACs gene cluster from strain HI1. The filled arrows denote the genes interrogated by the blast and are previously shown to be necessary for MACs function. The red filled arrow represents the macB gene, which is knocked out to create the nonfunctional MACs (Shikuma et al., 2014) strain used in the biofilm metamorphosis assays. (B) Maximum likelihood phylogeny of 19 sequenced P. luteoviolacea genomes including Pseudoalteromonas sp. ATCC 29581 as an outgroup. The bootstrap values represent 100 re-samples. The banded boxes indicate highly conserved subgroups for which symbol representations apply throughout the group. The blue pentagon denotes strains that produce TBP. Hollow pentagons represent strains that encode some genes in the bmp gene cluster, but do not experimentally produce halogenated compounds. Purple arrows indicate the presence of MACs genes (macB, macS, macT, and mif1).
