Significance
The intra-S phase checkpoint is essential to stabilize stalling/stalled replication forks. However, the mechanism underlying checkpoint regulation is poorly understood; two key questions remain to be answered: 1) why are stalling forks unstable when checkpoint is deficient, and 2) which protein(s) is the critical target of checkpoint regulation? Here, we demonstrate that CMG replicative helicase becomes uncoupled from stalled/blocked DNA polymerases in checkpoint-deficient cds1Chk2△ cells, likely reflecting physical separation of the helicase and polymerases. This uncoupling is prevented by the Cds1Chk2-mediated phosphorylation of Cdc45. We propose that replication elongation is directly regulated in response to replication fork stalling via phosphorylation of Cdc45, which significantly reduces CMG helicase processivity, in order to prevent helicase–polymerase uncoupling and promote replication fork stability.
Keywords: replication fork stability, checkpoint, CMG complex
Abstract
DNA replication is dramatically slowed down under replication stress. The regulation of replication speed is a conserved response in eukaryotes and, in fission yeast, requires the checkpoint kinases Rad3ATR and Cds1Chk2. However, the underlying mechanism of this checkpoint regulation remains unresolved. Here, we report that the Rad3ATR-Cds1Chk2 checkpoint directly targets the Cdc45-MCM-GINS (CMG) replicative helicase under replication stress. When replication forks stall, the Cds1Chk2 kinase directly phosphorylates Cdc45 on the S275, S322, and S397 residues, which significantly reduces CMG helicase activity. Furthermore, in cds1Chk2-mutated cells, the CMG helicase and DNA polymerases are physically separated, potentially disrupting replisomes and collapsing replication forks. This study demonstrates that the intra-S phase checkpoint directly regulates replication elongation, reduces CMG helicase processivity, prevents CMG helicase delinking from DNA polymerases, and therefore helps preserve the integrity of stalled replisomes and replication forks.
Eukaryotic DNA replisomes must overcome an extensive array of replication barriers to complete genomic DNA replication (1). These barriers include a great number of potential DNA secondary structures (e.g., G4, hairpins, triplexes), numerous proteins that tightly bind to DNA, various DNA lesions, and collisions with transcription apparatus (2–8). Most cancer cells also need to contend with dysregulated cell cycle progression that results in replication stress (9, 10). The cause of oncogene-induced replication stress is not fully understood but includes altered origin firing, increased replication–transcription clashes (11), misregulation of ribonucleotide reductase, and exhaustion of deoxyribonucleotide triphosphates (dNTPs) (12). All of these can result in the stalling of replication forks, which potentially causes fork instability, gross genomic alterations, and cancer (5, 13–16).
Stalling replication forks are unstable, and the intra-S phase checkpoint is absolutely essential for preventing such stalling forks from collapsing (5, 17–19). However, despite extensive studies in the past two decades, the reasons why stalling forks are unstable has not been fully defined. It is also not known precisely how the checkpoint acts to stabilize stalling forks. Previous studies have shown the formation of reversed forks and reported the dissociation of replication factors from stalled forks in checkpoint-deficient cells (17, 19–21). However, it remains to be clarified whether fork reversal and the dissociation of replication factors from stalling forks are a cause or a consequence of fork collapse. Most likely, fork collapse is a multistep process during which fork reversal and/or replication factor dissociation lead to increasingly severe fork damage until irreversible fork collapse occurs. In contrast to the findings of Cobb and Lucca (20, 21), who reported that DNA polymerases are lost from sites of replication stalling in budding yeast, more recent work from De Piccoli et al. (22) did not detect the dissociation of specific replication factors from stalling forks. The reasons for these contradictory data are unclear but may be related to the duration for which forks were stalled, how the replication proteins were isolated by chromatin immunoprecipitation (ChIP), and the sensitivity of the assays used to measure the global changes of specific replication factors within replisomes. For example, a 10 to 20% difference in a Western blotting assay may not appear very striking, but a collapse rate of 10 to 20% of the total replication forks would indeed be severe and may possibly be beyond cells capability to fix and thus complete DNA replication.
It has been reported that the Mrc1/Claspin protein forms a fork protection complex with Timeless/Tof1 and Tipin/Csm3 (Swi1 and Swi3 in Schizosaccharomyces pombe) and that this plays important roles in stabilizing stalling forks (23, 24). Mrc1 also plays a critical role in checkpoint activation and in the physical association between the replicative helicase and DNA polymerase ε (25–27). Thus, the requirement of Mrc1 for the stability of stalling forks is likely to be related to both its role in the checkpoint response and its function in the physical association of the CMG replicative helicase and DNA polymerase ε at replication forks. Besides the fork protection complex, the checkpoint also regulates several enzymes, preventing them from damaging stalling forks (28–30). In addition, in response to replication stress, an important aspect of checkpoint regulation is to inhibit late origin firing (31–35), which will reduce the number of replication forks encountering replication stress, control potential DNA damage, and therefore increase cell survival rate.
Furthermore, it was reported that, in budding yeast, the rate of DNA replication was dramatically slowed down in response to replication stress caused by DNA alkylation, and this was dependent on the checkpoint kinases Mec1 and Rad53 (36), indicating that replication slowing is an active process. In fission yeast checkpoint-deficient cells (cds1Chk2△ or mrc1△), DNA synthesis is prolonged in the presence of HU (hydroxyurea treatment) and remains dependent on the MCM helicase, and HU causes increasing fork collapse and DNA damage (37). Dna2, an essential flap endonuclease for processing Okazaki fragments in replication forks, is regulated by Cds1Chk2 for preventing fork reversal in response to replication fork stalling (17). Taken together, these studies suggest that checkpoint may directly target the replisome and regulate the replication elongation step.
When replication is experimentally impeded by HU (a common tool for studying replication fork stalling that inhibits ribonucleotide reductase, thus impeding DNA polymerization), the ATR-dependent intra-S phase checkpoint stabilizes stalling replication forks such that fork pausing is transient and can be relieved when the HU is removed. Furthermore, when the checkpoint is deficient, stalling forks rapidly collapse, leading to irreparably dysfunctional replication (18, 38), pathological reversed forks (17, 19), DNA breaks (5), and unscheduled recombination (13, 39). However, due to the fact that only a limited number of checkpoint targets have been identified and their regulation mechanisms elucidated, two key questions remain to be answered (14, 17, 40, 41): 1) Why are stalling forks unstable when the checkpoint is deficient, and 2) which protein factor or factors are the critical target of regulation by checkpoint?
In fission yeast, the Cds1Chk2 kinase defines the major intra-S phase branch of the Rad3ATR checkpoint. In mammalian cells, this ATR-dependent function is performed mainly by Chk1, a functional homolog of yeast Chk2. Mammalian Chk2 responds to ATM activation in response to DSBs (42). Fission yeast cds1Chk2△ cells are hypersensitive to HU due to rapid fork collapse and global replication failure (17, 43). We therefore searched for intra-S phase checkpoint targets by screening for spontaneous genetic mutations that conferred HU resistance to fission yeast cds1Chk2△ cells. Our rationale was that, if a mutation located in a checkpoint target partially mimics the intra-S phase checkpoint regulation, it will render checkpoint-deficient cells more HU resistant. We identified an R319I mutation in cdc45 (cdc45-R319I) encoding a subunit of the CMG (Cdc45-MCM-GINS) replicative helicase. cdc45-R319I rescued the 2.5 mM HU sensitivity of cds1Chk2△ cells by ∼600- to 3,000-fold, suggesting that Cdc45 may be a crucial target of the checkpoint response to replication stress. Subsequently, we demonstrate that Cdc45 is directly targeted by the Rad3ATR-Cds1Chk2 checkpoint pathway: when replication forks stall in the presence of HU, Cds1Chk2 directly phosphorylates Cdc45 on S275, S322, and S397. This Cds1Chk2-mediated Cdc45 phosphorylation dramatically reduces CMG helicase activity, resulting in the slowing of replication elongation.
Using a ChIP assay (44), we further demonstrate that the CMG replicative helicase becomes uncoupled from stalled/blocked DNA polymerases in checkpoint-deficient cds1Chk2△ cells, likely reflecting the physical separation of the helicase and polymerases. This uncoupling is prevented by the Cds1Chk2-mediated Cdc45 phosphorylation. Thus, we propose that replication elongation is directly regulated in response to replication fork stalling via the phosphorylation of Cdc45, which reduces CMG helicase processivity in order to help prevent helicase–polymerase uncoupling and promote replication fork stability.
Results
Cds1Chk2 Targets the Replicative Helicase CMG Complex.
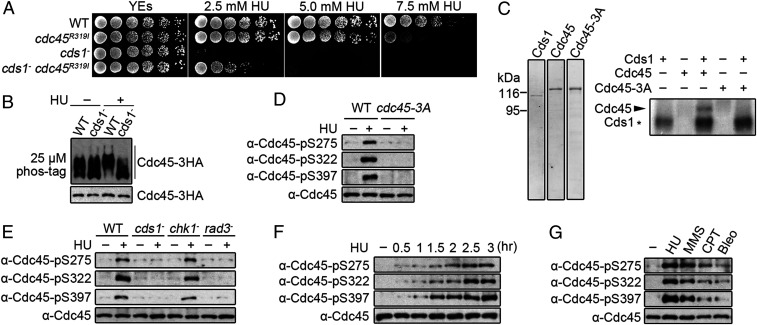
To explore how the intra-S phase checkpoint pathway regulates replication forks, we established a genetic screen to identify mutations that may mimic checkpoint regulation. We reasoned that mutants which render cds1Chk2△ cells resistance to replication fork stalling by HU treatment (45) may be located in checkpoint targets and may act by mimicking checkpoint regulation. cds1Chk2△ cells were plated on rich media agar containing 5 mM HU, and colonies emerging after several days of incubation (SI Appendix, Fig. S1 A and B) were isolated and confirmed for HU resistance (SI Appendix, Fig. S1C). Genomic DNA was then extracted and subjected to deep sequencing. The mutations identified were individually reintroduced into a cds1Chk2△ background to establish if they restored HU resistance. A cdc45-R319I mutation was identified that dramatically suppressed the HU sensitivity of cds1Chk2△ cells (Fig. 1A). When assayed in a cds1+ background, cells harboring the cdc45-R319I mutation were not sensitive to 2.5 mM HU and exhibited little sensitivity to 5.0 mM. At higher concentrations, sensitivity became more pronounced (Fig. 1A). The cdc45-R319I mutation is recessive: introducing a second wild-type copy of cdc45 by integrating cdc45+ into the ura4 locus prevented the rescue of HU sensitivity of cds1Chk2△ cells by cdc45-R319I (SI Appendix, Fig. S1D).
Fig. 1.
Cds1Chk2 phosphorylates Cdc45 on S275, S322, and S397 when replication forks stall. (A) A fivefold serial dilution of the indicated stains grown with the designated concentration of HU. (B) Cdc45 phosphorylation in response to HU. (Top) Phos-tag sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). (Bottom) SDS-PAGE. HA-tagged Cdc45 was detected by α-HA. (C, Left) Purified Cds1, Cdc45-MBP-tag, and Cdc45-S275A-S322A-S397A (S3A). (Right) Autoradiograph of purified Cdc45 following in vitro phosphorylation by Cds1Chk2. (D) Detection of Cdc45 phospho-S275, -S322, and -S397 in vivo using phospho-specific antibodies. (E) Phosphorylation of Cdc45 S275, S322, and S397 in the indicated strains. (F) Time course of S275, S322, and S397 phosphorylation following HU treatment. (G) Cdc45 phosphorylation in response to the indicated genotoxic agents.
Following HU treatment, we observed that Cdc45 displayed a cds1Chk2-dependent mobility shift on sodium dodecyl sulphate–polyacrylamide gel electrophoresis gels containing 25 μM Phos-tag, indicating that Cdc45 is phosphorylated in response to fork stalling (Fig. 1B). Mass spectroscopy (MS) determined that the S275, S322, and S397 sites of Cdc45 are phosphorylated (SI Appendix, Fig. S1E). In vitro, Cds1Chk2 directly phosphorylated Cdc45-MBP-tag (Fig. 1C), and no Cdc45 phosphorylation was observed when the S275, S322, and S397 residues were mutated to alanine, confirming that S275, S322, and S397 were also in vitro Cds1Chk2 targets (Fig. 1C). Together with the analysis of the cdc45-R319I mutant, the Cds1Chk2-and replication stress–dependent phosphorylation of Cdc45 strongly suggests that Cdc45 is a checkpoint target in response to fork stalling. We thus developed our subsequent studies to confirm the Cds1Chk2-dependent phosphorylation of Cdc45 and examine its physiological function and mechanism in stabilizing stalling replication forks. The cdc45-R319I mutant was not analyzed further since it is a random untargeted mutation, although it did provide the clue that Cdc45 may be regulated in response to replication stress.
Phospho-specific antibodies were prepared against each of the three sites (SI Appendix, Fig. S1F) and used to confirm that the in vivo phosphorylation of Cdc45 on S275, S322, and S397 depended on presence of HU, Cds1Chk2, and Rad3ATR but not on Chk1 (Fig. 1 D and E). Furthermore, all three serine residues were concurrently phosphorylated over time upon HU exposure (Fig. 1F), and the phosphorylation was elicited more avidly by agents that interfere with replicative polymerases (HU or methyl methanesulfonate [MMS]) when compared to agents such as camptothecin (CPT) that predominantly block replicative helicase progression or bleomycin (Bleo) that induces DNA breaks (Fig. 1G). Taken together, these results demonstrate that Cdc45 is phosphorylated on S275, S322, and S397 by Cds1Chk2 in response to replication fork stalling by agents that interfere with DNA synthesis.
S275, S322, and S397 are all located within the CID domain where Cdc45 interacts with MCM and GINS. S275 and S322 but not S397 lie within a consensus motif for Cds1Chk2 in S. pombe (SI Appendix, Fig. S1G). The Cds1Chk2 consensus motif is I/LxR/KxxS/T with some flexibility (46, 47). S322 and S397 are highly conserved from yeast to human cells. S275 is not conserved, but human Cdc45 has threonine and serine residues (three and seven residues, respectively) from the site corresponding to S275 of S. pombe Cdc45. These sites are conserved from Xenopus laevis to Homo sapiens and both are within a loose Chk2 consensus motif. Thus, Cds1Chk2-mediated Cdc45 phosphorylation on S275, S322, and S397 may be conserved in eukaryotes.
cdc45-3A and cdc45-3D Mutations Stabilize Stalling/Stalled Replication Forks in Checkpoint-Deficient Cells.
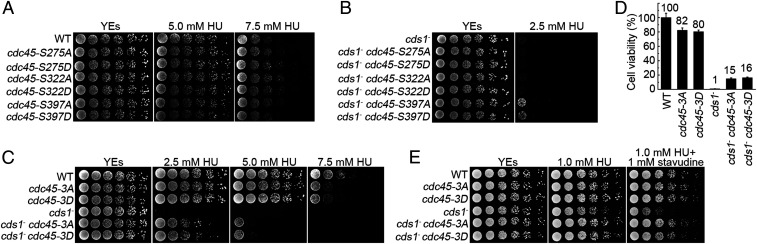
To examine the role of Cds1Chk2-mediated Cdc45 phosphorylation in stabilizing stalling forks, S275, S322, and S397 were mutated individually or all together, either to alanine (phospho deficient) or aspartic acid (potential phospho mimic). Individual mutations displayed similar growth rates, and HU sensitivities compared to wild-type (wt) cells (Fig. 2A). Mutations at S275 or S322 did not rescue the HU sensitivity of cds1Chk2△ cells, whereas cdc45-S397A and cdc451-S397D modestly reduced the HU sensitivity only at low HU concentrations (Fig. 2B). However, the triple cdc45-3A and cdc45-3D mutations significantly rescued the HU sensitivity of cds1Chk2△ cells (Fig. 2C). From these spot-test assays, in which cells are grown under a chronic exposure to HU, we estimated an ∼600- to 3,000-fold increase in survival when compared to cds1Chk2△ cdc45+ cells. A clonogenic survival assay, in which cells were transiently exposed to 12.5 mM HU for 3 h, led to increased cell survival when compared to the cds1Chk2△ single mutant of ∼15- to 20-fold (Fig. 2D). Both the cdc45-3A and cdc45-3D mutants showed a similar level of modest sensitivity to high HU concentration (7.5 mM) in a cds1+ background when compared to cdc45+ cells (Fig. 2 C and D). The basically equivalent rescue effect of the cdc45-3A and cdc45-3D mutations on the HU sensitivity of cds1Chk2△ cells suggests that both alleles alter the biochemical property of the CMG helicase in a similar way (see Fig. 5).
Fig. 2.
Cdc45-3A/3D mutations significantly rescue the HU sensitivity of cds1Chk2△ cells. (A) Fivefold serial dilution assays of the indicated single cdc45 phospho-site mutant strains grown with the indicated HU concentrations. WT = cdc45+ control. (B) Equivalent serial dilution assay in a cds1Chk2△ genetic background. (C) Equivalent serial dilution assay of the indicated triple phospho-site cdc45 mutations in cds1+ (WT) and cds1∆ backgrounds. (D) The relative cell viability of the indicated strains after treatment with 12.5 mM HU for 3 h. The data are presented as the mean ± SD. (E) Serial dilution assay of the indicated triple phospho-site cdc45 mutations in cds1+ (WT) and cds1∆ backgrounds in the presence of the dideoxynucleoside analog stavudine.
Fig. 5.
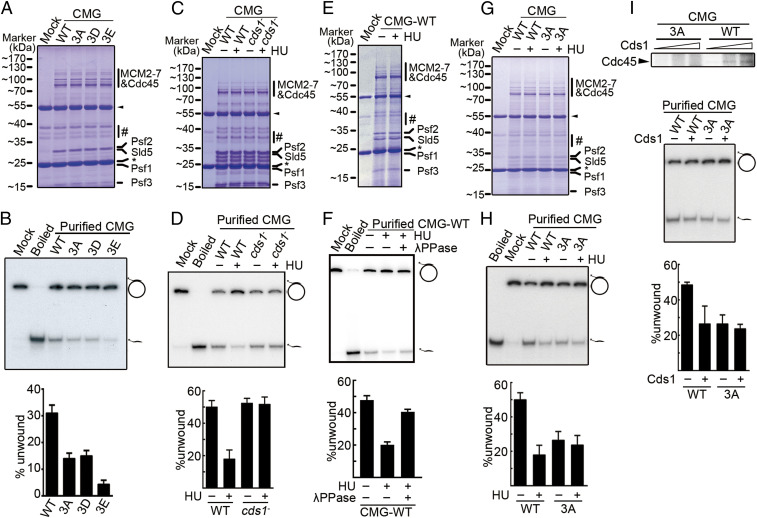
Cds1Chk2-mediated phosphorylation of Cdc45 regulates CMG helicase activity. (A) Coomassie-stained CMG complexes purified from cdc45+, cdc45-3A, cdc45-3D, and cdc45-3E (CMG, CMG-3A, CMG-3D, and CMG-3E, respectively) growing without HU treatment. Heavy and light antibody chains are indicated by a black arrow and star, respectively. #: contaminants from HA antibody-conjugated resins. (B, Top) Autoradiograph reporting the helicase activity of CMG and CMG-3A/-3D/3E. The position of the substrate (32P-labeled oligonucleotide annealed to a M13 circular ssDNA) and displaced oligonucleotide were indicated. (Bottom) Quantification of three independent experiments. (C, E, and G) Indicated CMG complexes purified from untreated or HU-treated cells as in A. (D, F, and H) Autoradiograph and quantification of the helicase activity of purified CMG complexes shown in C, E, and G (see B for details). The presence of λPPase is indicated in F. (I, Top) Cds1-dependent phosphorylation of the Cdc45 subunit of the indicated CMG complexes in vitro. (Bottom) Autoradiograph and quantification of the helicase activity of the indicated CMG complexes with and without in vitro Cds1 treatment (see B for details). The data in B, D, F, H, and I are presented as the mean ± SD.
The critical role of the Cdc45 residues S275, S322, and S397 for the biochemical property of CMG helicase was further examined. These three serine residues were mutated to either glutamate (cdc45-3E, bearing a larger residual group compared to aspartate), threonine (cdc45-3T, similar to serine except having an extra methyl group), cysteine (cdc45-3C, similar to serine but with thiol replacing hydroxyl group), lysine (cdc45-3K, positive charge), asparagine (cdc45-3N, possessing a chemically polar residual group), glycine (similar to alanine but possessing a smaller residual group), and valine (hydrophobic residual group). These new mutants were all examined for their growth in the presence or absence of HU and for their rescue effect on the HU sensitivity of cds1Chk2△ cells. As shown in SI Appendix, Fig. S1H, these mutants, with the exception of cdc45-3E, which exhibited a severe growth defect, grew similarly well compared to cdc45+ cells in normal medium (HU absent). The viable mutants, with the exception of cdc45-3T, showed apparent growth sensitivity to 5.0 or 7.5 mM HU (SI Appendix, Fig. S1H), and the level of the HU sensitivity among the mutants was slightly different, with cdc45-3A, cdc45-3D, cdc45-3C, cdc45-3K, and cdc45-3V being more sensitive to HU than cdc45-3N and cdc45-3G (SI Appendix, Fig. S1H). The cdc45-3E mutant did not grow in the presence of 5.0 or 7.5 mM HU (SI Appendix, Fig. S1H). With the exception of cdc45-3E (which could not be measured due to a severe growth defect), all the cdc45 mutants rescued the HU sensitivity of cds1Chk2△ cells. The levels of rescue were different: cdc45-3A, cdc45-3D, and cdc45-3C > cdc45-3K, cdc45-3V > cdc45-3N, and cdc45-3G > cdc45-3T (SI Appendix, Figs. S1I and S5D). These results suggest that the S275, S322, and S397 residues in Cdc45 are critical for preserving the normal activity/function of CMG helicase; a slight difference either in spatial conformation (threonine contains an extra methyl group compared to serine) or in chemical polarity (i.e., cysteine versus serine) alters the properties of the CMG helicase, rendering cds1Chk2△ cells more resistant to HU. The following studies focused on the cdc45-3A and cdc45-3D mutations in order to gain further insight into the mechanism of how the checkpoint regulates the CMG helicase to stabilize stalling/stalled replication forks.
HU stalls replication forks by limiting the availability of dNTPs, thus limiting the processivity of the replicative polymerases. To establish if Cdc45 phosphorylation site mutants could rescue cds1Chk2△ cell sensitivity to other polymerase stalling agents, we examined the response to stavudine. Stavudine is a dideoxyribonucleoside analog that is converted to active stavudine triphosphate by cellular kinases in vivo and subsequently incorporated into the leading and lagging strands. The incorporation of stavudine blocks further incorporation of downstream nucleotides and thus stalls replication forks (48). In order to enhance the chance of stavudine incorporation, the cellular concentration of dNTPs was reduced by including 1 mM HU in medium. When treated with 1 mM HU, all strains showed similar growth rates, except cds1Chk2△ cells that showed a very modest sensitivity (Fig. 2 E, Middle). When both 1 mM of stavudine and 1 mM of HU were present, wt, cdc45-3A, and cdc45-3D strains still displayed a similar growth rate, but cds1Chk2△ cells were ∼25-fold more sensitive when compared to wt cells (Fig. 2 E, Right). However, the cds1Chk2△ cdc45-3A and cds1Chk2△ cdc45-3D strains showed an ∼25-fold of reduction in stavudine sensitivity when compared to cds1Chk2△ cells. These data further confirm that the cdc45-3A and cdc45-3D mutations significantly reduce the sensitivity of cds1Chk2△ cells to reagents that stall replication forks.
cdc45-3A and cdc45-3D Mutations Reduce Collapsing Rate of Stalled Replication Forks.
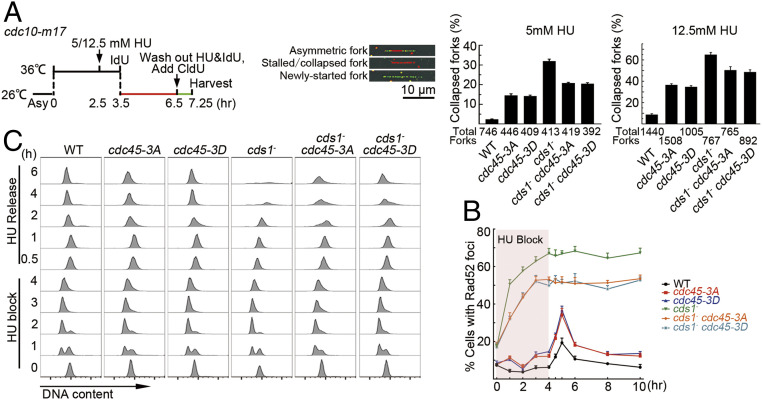
To establish how the cdc45-3A or cdc45-3D mutations influence the stabilization of stalling forks, we used dual-color DNA combing to assess the stability of DNA replication forks upon 5 or 12.5 mM HU treatment (Fig. 3 A, Left). Examples of fibers with asymmetric, stalled, or collapsed and newly started forks are shown (Fig. 3 A, Middle). The percentage of permanently arrested forks (IdU fibers without CldU tracks at one or both ends) increased in cdc45-3A and cdc45-3D cells when compared to cdc45+ (5 mM HU: 14.5, 14, and 2.5%; 12.5 mM HU: 36, 35, and 9%, respectively). As expected, in cds1Chk2△ (checkpoint-deficient) cells, the percentage of arrested forks unable to resume replication was higher, at ∼31 and 65% with 5 or 12.5 mM HU treatment, respectively (Fig. 3 A, Right). Under 5 or 12.5 mM HU treatment for 4 h, both cdc45-3A and cdc45-3D mutations reduced the frequency of permanently arrested forks in the cds1Chk2△ background by ∼34 ([31 − 21]/31) or 25% ([65 − 49]/65) (Fig. 3 A, Right), indicating that even under a short time of HU treatment, a considerable number of replication forks collapsed in cds1Chk2△ cells, but the cdc45-3A and cdc45-3D mutations significantly reduced the rate of fork collapsing under Cds1Chk2 deficiency. In addition, the cdc45-3A and cdc45-3D cells had a higher rate of fork collapsing than cdc45+ cells, indicating that cdc45-3A and cdc45-3D mutations do not fully mimic Cds1Chk2 regulation to Cdc45 (see Fig. 5H, the level of helicase activity).
Fig. 3.
Cdc45-3A/3D mutations stabilize stalled replication forks. (A) Dual labeling combing to assess the rate of stalled fork collapse. (Left) Experimental scheme. (Middle) Examples of fiber staining. (Right) Percentage of collapsed forks in the indicated genetic backgrounds calculated as the number of IdU-only DNA tracts divided by the total tracts. (B) Time course of the percentage of cells showing Rad52 foci during HU treatment (12.5 mM) for 4 h (pink box) and subsequent release. (C) FACS analysis of cells from B. The data in A and B are presented as the mean ± SD.
Fluorescence-activated cell sorting (FACS) analyses were performed to those cells that were analyzed in Fig. 3A. As shown in SI Appendix, Fig. S2A, these cells were arrested in the G1 phase after 3.5 h at 36 °C. After the cultures were shifted to a permissive temperature of 26 °C with or without HU, the wt and cds1Chk2△ cells progressed nicely to the G2 phase with a DNA content of 2C in the absence of HU (the peaks are slightly skewed to the right 3 h after cell release compared to the 2C peaks before arrest [“0” h] because the cells became larger during arrest) (SI Appendix, Fig. S2 A, Top and Bottom). However, a similar level of DNA content of ∼1C (the peaks are slightly skewed to the right due to a larger cell size) was detected among wt, cds1Chk2△, cdc45-3A, cdc45-3D, cdc45-R319I, cds1Chk2△ cdc45-3A, cds1Chk2△ cdc45-3D, and cds1Chk2△ cdc45-R319I cells (SI Appendix, Fig. S2 A, Top and Bottom). These results suggest that the forks without CldU incorporation represent stalled or collapsed forks rather than resulting from the merging of two neighboring forks.
Fork collapse results in DNA lesions that recruit the Rad52 recombination protein to repair the broken forks. During repair, Rad52 forms a focus. Thus, we monitored Rad52 foci as an indirect readout of fork collapse in cells treated with HU and then released into fresh media (SI Appendix, Fig. S2B). Under unperturbed growth condition (−HU) ∼8% of wt cells and ∼18% of cds1Chk2△ cells showed Rad52 foci (Fig. 3B). cdc45-3A and cdc45-3D were similar to wt, whereas the cds1Chk2△ cdc45-3A and cds1Chk2△ cdc45-3D double mutant strains were similar to the cds1Chk2△ single mutant (Fig. 3B). As previously reported (37), during 4 h of HU treatment, the percentage of wt cells exhibiting Rad52 foci did not increase, whereas the percentage of the Rad52 foci positive cds1Chk2△ mutant increased to ∼67%. Consistent with the cdc45-3A and cdc45-3D mutations partially suppressing the cds1Chk2△ mutant sensitivity to HU, the cds1Chk2△ cdc45-3A and cds1Chk2△ cdc45-3D double mutants displayed a less dramatic increase in Rad52 foci positive cells at ∼50%. The single cdc45-3A and cdc45-3D mutants showed only a modest increase (∼14%). Upon release from HU, wt and the single cdc45-3A and cdc45-3D mutants showed a transient pulse of Rad52 foci positive cells, whereas the percentage of Rad52 foci positive cells observed after the release from HU arrest did not change in cds1Chk2△ or the cds1Chk2△ cdc45-3A and cds1Chk2△ cdc45-3D double mutant cultures (Fig. 3B). Flow cytometry assays showed that the cdc45-3A and cdc45-3D mutations increased the ability of cds1Chk2△ cells to re-enter the cell cycle after 4 h of HU treatment (Fig. 3C). These results are consistent with our interpretation that the cdc45-3A and cdc45-3D mutations reduce the rate of collapse of stalling forks in cds1Chk2△ cells.
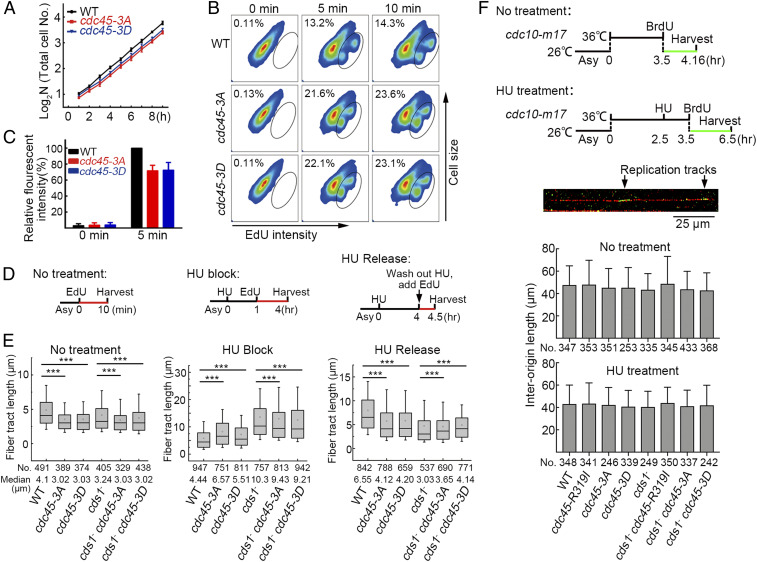
Checkpoint Regulation and the cdc45-3A and cdc45-3D Alleles Decrease Fork Speed.
cdc45-3A and cdc45-3D cultures grow slightly more slowly than cdc45+ (Fig. 4A). Pulse labeling of asynchronous cultures with EdU demonstrated that the number of S phase cells in cdc45-3A and cdc45-3D increased by ∼65% compared to cdc45+ cultures (Fig. 4B). Consistent with an extended S phase, the average amount of EdU incorporated per cell in cdc45-3A and cdc45-3D was decreased by ∼30% (Fig. 4C). This suggests that the reduced growth rate results from an extended S phase, probably because of slower replication fork progression. To address this, DNA combing was used to examine fork speed in cdc45+ and appropriate mutant cells during the unperturbed S phase, during HU treatment, and after release from HU arrest (schematic, Fig. 4D). SI Appendix, Fig. S3 A and B show examples of the labeled DNA fibers and the distributions of their lengths. During the unperturbed S phase (10 min EdU incorporation), the median tract length for cdc45+ cells was 4.1 μm (Fig. 4 E, Left), translating to ∼14∼21 nucleotides per second (nt/s), based on ∼2 to 3 kb/per μm of DNA fiber (49). cdc45-3A and cdc45-3D cells displayed a reduced fork speed (3.02 and 3.03 μm, respectively: 10∼15 nt/s). This indicates that Cdc45-3A and Cdc45-3D reduce the activity/processivity of the CMG helicase. Fork speed was also decreased in the cds1Chk2△, cds1Chk2△ cdc45-3A, and cds1Chk2△ cdc45-3D cells. While the explanation for a slower fork speed in the cds1Chk2△ cells is unknown, it is possible that a basal level of checkpoint regulation is required to promote replication fork movement through difficult-to-replicate loci (5).
Fig. 4.
Fork speed correlates with HU survival. (A) Growth curves of the indicated strains. (B) The percentage of cells in S phase measured after 5 and 10 min EdU pulses by FACS in the indicated strains. EdU-labeled (S phase) cells are encircled. (C) The relative fluorescent intensity of EdU-stained cells. (D) Experimental scheme for measuring fork speed by DNA combing. (E) The medium length of EdU-labeled DNA fibers in the WT and indicated mutant strains under normal cell growth conditions in the presence of HU and after release from HU treatment. The numbers of measured DNA fibers are indicated. Representative images are shown in SI Appendix, Fig. S3A. (F) The measurement of interorigin distance in wt, cds1−, cdc45-3A, cdc45-3D, cdc45-R319I, cds1− cdc45-3A, cds1− cdc45-3D, and cds1− cdc45-R319I cells in the presence or absence of HU. (Top) Experimental scheme. (Bottom) Average interorigin distance in the indicated strains. The data in A, C, and F are presented as the mean value ± SD. The data in E is presented as a box plot with an interquartile range of 25 to 75%, the median value (line), mean value (small hollow square), limited inferior <5%, and limited superior >95%. The Mann–Whitney U test was applied for statistical analysis. ***P < 0.001.
To examine fork speed in the presence of HU, the cultures were first incubated with HU for 1 h. Then, EdU was added, and the cultures incubated for an additional 3 h. As shown in Fig. 4 E, Middle, fork speed in cdc45+ cells dramatically decreased; a tract length of 5 μm equates to a speed of 1 nt/s. A less dramatic decrease (∼2 nt/s) was observed for cds1Chk2△ cells. cdc45-3A and cdc45-3D single mutant cells displayed a fork speed appreciably above cdc45+ cells but substantially below that of cds1Chk2△ cells. Importantly, the double mutant cds1Chk2△ cdc45-3A and the cds1Chk2△ cdc45-3D cells exhibited a fork speed that was appreciably below cds1Chk2△ cells but still substantially above that of cdc45+ cells. These data are again consistent with Cdc45-3A and Cdc45-3D reducing the activity/processivity of the CMG helicase, therefore partially mimicking the regulation of Cdc45 by Cds1Chk2. Following the release from HU arrest, replication forks quickly recovered in cdc45+ cells. Forks recovered to a much lesser extent in cds1Chk2△ cells. As expected, cdc45-3A and cdc45-3D mutants recovered less well than cdc45+ cells but better than cds1Chk2△ cells. Importantly, cds1Chk2△ cdc45-3A and cds1Chk2△ cdc45-3D double mutant cells recovered appreciably better than cds1Chk2△ cells, although substantially less well than cdc45+ cells (Fig. 4 E, Right). Thus, the recovery of replication following HU treatment and the extent of changes in replication fork speed correlate with HU sensitivity, suggesting fork progression is a critical target of the intra-S phase checkpoint regulation in response to replication fork stalling.
Changes in fork speed have been correlated with changes in origin firing (50). To establish if wt, cds1Chk2△, cdc45-3A, cdc45-3D, cdc45-R319I, cds1Chk2△ cdc45-3A, cds1Chk2△ cdc45-3D, and cds1Chk2△ cdc45-R319I cultures display similar efficiencies of DNA replication initiation, the “start” cell cycle control gene, cdc10+, was replaced by the temperature-sensitive cdc10-m17 allele. Cells were arrested at the G1 phase and subsequently released into a synchronous S phase in the presence of BrdU for 40 min (−HU) or 3 h (+HU). DNA combing was then performed and interorigin distances estimated from BrdU staining (51). As shown in Fig. 4 F, Bottom, no matter whether HU was present or absent, the average distance between two neighboring initiation sites among these cells was equivalent. Thus, the efficiency of replication initiation is not affected by the cds1Chk2△, cdc45-3A, cdc45-3D, cdc45-R319I, or the cells bearing cds1Chk2△ combined with the cdc45 mutant. These results are consistent with that the fission yeast S. pombe cells have only eight late origins (1 to 2% of total origins) (52) and that only a few origins in S. pombe are affected by checkpoint regulation in response to replication stress (53).
Cds1Chk2-Mediated Cdc45 Phosphorylation Inhibits CMG Helicase Activity.
To establish if Cds1Chk2-mediated Cdc45 phosphorylation reduces CMG helicase activity, we assayed the helicase activity of CMG isolated from the chromatin of unperturbed wt and mutant cells and from cells experiencing DNA replication stress. Under unperturbed growth conditions, the helicase activities of CMG-3A and CMG-3D were reduced by ∼50%. A second potential phosphor-mimic mutant, cdc45-3E, grew very poorly (SI Appendix, Fig. S4 A–C) and displayed an 85% reduction in helicase activity (Fig. 5 A and B). Under replication stress (the presence of HU), the helicase activity of CMG from wt cells was reduced by ∼70% but remained unchanged when isolated from cds1Chk2△ cells (Fig. 5 C and D). In support of this result, the helicase activity of the CMG complex isolated from HU-treated cells was examined in the presence or absence of λPPase. As shown in Fig. 5 E and F, the CMG helicase activity was dramatically reduced in the HU-treated cells, but the helicase activity was nearly recovered to the level of normal CMG helicase activity. This result indicates that the CMG activity is greatly reduced in stalling replisomes, and this reduction appears solely dependent upon Cds1Chk2 checkpoint regulation. Consistent with Cds1Chk2 activity inhibiting CMG via phosphorylation of Cdc45 S275, S322, and S397, when CMG and CMG-3A were isolated in parallel from cells either treated, or not, with HU (Fig. 5 G and H), the helicase activity was reduced by ∼75% for CMG, whereas the activity of CMG-3A, which is already compromised, was not significantly further reduced. This is consistent with Cdc45-3A being largely insensitive to Cds1Chk2 regulation (Fig. 1C) and suggests that there are no further major modifications on Cdc45 or other subunits of CMG complex by the intra-S phase checkpoint that reduce CMG helicase activity.
When CMG or CMG-3A were treated in vitro by Cds1Chk2, Cdc45, but not Cdc45-3A, was phosphorylated (Fig. 5 I, Top). The helicase activity of CMG was reduced by ∼50%, whereas there was no significant change in the activity of CMG-3A. The difference in the reduction of helicase activity (∼75% in vivo versus ∼50% in vitro) is probably because the level of Cds1-mediated phosphorylation of Cdc45 in the in vitro reaction did not reach to a maximum extent that is reached in vivo. No detectable changes in the level of Cdc45, formation of CMG complex, or affinity of CMG to chromatin DNA were observed between cdc45+ and cdc45-3A cells, either with or without HU treatment (SI Appendix, Fig. S5 A–C), which is consistent with that the cdc45-3A or cdc45-3D cells grew relatively normally in the absence of exogenous replication stress. However, the S phase (∼10% of the cell division cycle) of these mutant cells is lengthened by ∼65% (Fig. 4 A and B). And this suggests that the conformation of the mutant CMG complex should be changed in some points, which results in a reduced helicase activity (Fig. 5 A and B). Taken together, our data suggests that Cds1Chk2-mediated Cdc45 phosphorylation on S275, S322, and S397 reduces CMG helicase activity and slows fork speed in response of replication fork stalling.
Cds1Chk2 Prevents DNA Polymerases and CMG Helicase in Stalling Forks from Separation.
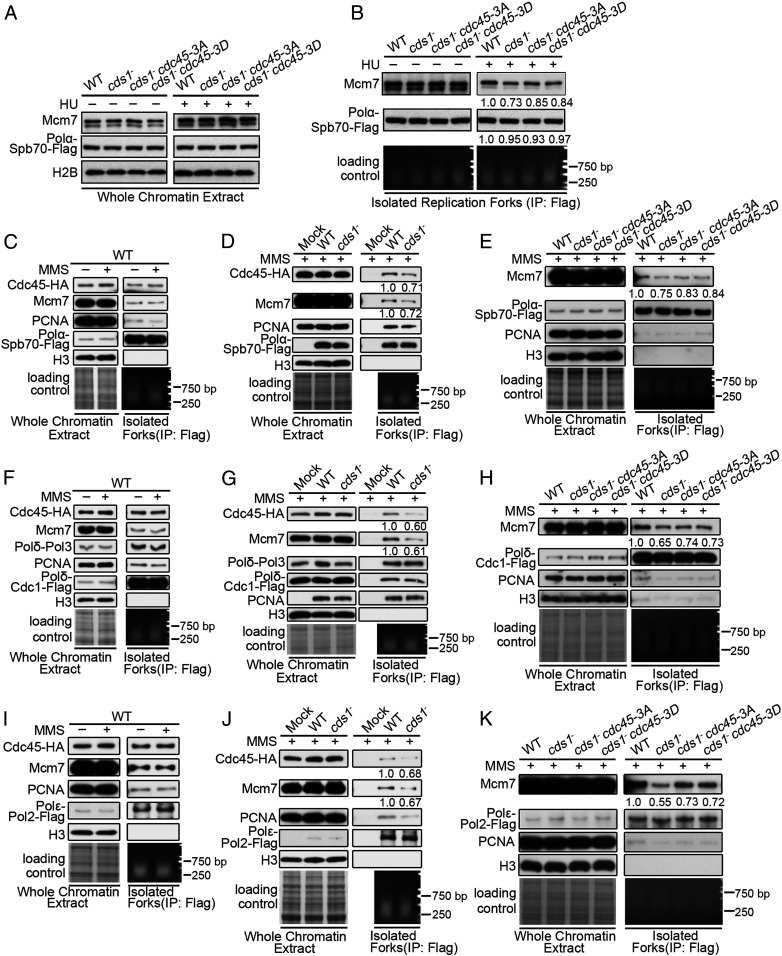
The Cds1Chk2-mediated inhibition of CMG helicase activity at stalling replication forks implies that a key mechanism in checkpoint regulation to preserve replisome integrity is to maintain the link between strand separation and DNA polymerization. Thus, we examined whether two core components, DNA polymerase α and the CMG helicase, are separated when replication forks are stalling. The levels of DNA polymerase α (Spb70 subunit) and MCM (Mcm7 subunit) were assessed in normal and stalling forks in wt and cds1Chk2△ cells. First, we observed that the levels of Spb70 (Pol-α subunit) and Mcm7 (CMG subunit) on chromatin were similar in the presence or absence of HU for wt, cds1Chk2△, cds1Chk2△ cdc45-3A, and cds1Chk2△ cdc45-3D cells (Fig. 6A). When replication forks were isolated from the chromatin by immunoprecipitation against DNA Pol-α (Spb70 subunit), the amount of the two subunits was equivalent between the four cultures when replication forks were not stalled (Fig. 6B). However, when the replication forks were isolated from cells treated with HU, the levels of Spb70 remained comparable between the cultures, but the level of Mcm7 decreased by ∼27, ∼15, and ∼16%, respectively, in cds1Chk2△, cds1Chk2△ cdc45-3A, and cds1Chk2△ cdc45-3D cells (Fig. 6B). A physical separation of replicative helicase and DNA polymerases collapses forks because a normal/functional replication fork requires both replicative helicase and DNA polymerases present in forks, and their actions must be strictly coupled. The above data indicate that ∼1/6 and 7 to 1/4 of forks collapsed after 3 h of HU treatment in the cds1Chk2△ cdc45-3A, cds1Chk2△ cdc45-3D, and cds1Chk2△ cells. These results suggest that the inhibition of CMG helicase activity is required to prevent the separation of the CMG replicative helicase and DNA polymerase α.
Fig. 6.
Cds1Chk2 prevents separation of DNA polymerases and CMG helicase in stalling replication forks. (A) The levels of Pol-α-Spb70 and Mcm7 on chromatin from the indicated strains in presence or absence of HU. (B) The amount of Mcm7 in stalling forks isolated from the indicated strains. (C–K) The levels of Mcm7, Cdc45, PCNA, Pol-α-Spb70 (C–E), Pol-δ-Cdc1 (F–H), and Pol-ε-Pol2 (I–K) on chromatin isolated from unperturbed or MMS-treated (forks stalling) cells of wt, cds1−, cds1− cdc45-3A, and cds1− cdc45-3D. Loading controls, Coomassie blue-stained gel, and isolated fork DNA are shown at the bottom of the panels.
The separation of CMG and DNA polymerases in the cds1Chk2△ cells was further examined in response to replication fork stalling due to MMS treatment (54), with replication forks isolated by immunoprecipitation against either DNA Pol-α, Pol-δ, or Pol-ε. The results are shown in Fig. 6 C–K. The total amount of Pol-α (Spb70), Pol-δ (Cdc1 and Pol3), Pol-ε (Pol2), PCNA, Mcm7, or Cdc45 on chromatin was equivalent between wt and cds1Chk2△ cells (Fig. 6 C–D, F, G, I, and J). In the isolated forks (immunoprecipitated with polymerase subunits), the amount of Spb70, Cdc1, Pol3, Pol2, or PCNA remained equal, and the amounts of Mcm7 and Cdc45 also remained equal in wt cells (Fig. 6 C, F, and I), but the amounts of Mcm7 and Cdc45 were reduced by ∼30 to 40% in the cds1Chk2△ cells, again consistent with the separation of helicase and DNA polymerases in stalling forks (Fig. 6 D, G, and J). As in the case of HU stalling replication forks, the cdc45-3A and cdc45-3D mutants, which partially mimic the checkpoint regulation on Cdc45, reduced the level of the CMG helicase and DNA polymerases separation (Fig. 6 E, H, and K). In addition, the physical separation of the replicative helicase CMG and DNA polymerases helicase is supposed to increase the amount of single-stranded DNA (ssDNA) on chromatin and potentially cause more DNA breaks. By measuring RPA foci, the cellular level of broken DNA, and the sensitivity of chromatin DNA to the ssDNA-specific endonuclease P1 in the HU-treated wt, cds1Chk2△, cds1Chk2△ cdc45-3A, and cds1Chk2△ cdc45-3D cells, the results show that RPA foci significantly increased in cds1Chk2△ cells, but Cdc45-3A and Cdc45-3D mutations noticeably reduced RPA foci in the cds1Chk2△ cells (SI Appendix, Fig. S6 A, B, and D). Consistent with more RPA foci, an ∼28% increase in the level of broken DNA was detected in the cds1− cells compared with wt, no matter whether chromatin DNA was treated or untreated by nuclease P1 (SI Appendix, Fig. S6C), suggesting that chromatin DNA in the cds1− cells is more vulnerable, probably due to more ssDNA. Taken together, these results indicate that the CMG helicase moves away from DNA polymerases or replication forks when replication forks are stalling in the absence of the intra-S phase checkpoint kinase Cds1Chk2 and that a reduction of CMG helicase activity (in the cdc45-3A/3D mutants) alleviates this separation.
Discussion
The catalytic centers in replicative DNA polymerases have an ultra-fine structure (55). These centers can accommodate normal A:T and G:C pairs but do not accommodate either A:C or G:T pairs, even though the three-dimensional difference between the A:T and A:C or the G:C and G:T pair is small. An ultra-fine catalytic center greatly increases the fidelity of DNA synthesis, with an error rate of ∼10−4 per base incorporated (56). However, it also means that DNA polymerases can be easily blocked, causing fork stalling. Base modifications, such as pyrimidine dimerization, base loss, base alkylation, and base oxidation, can block DNA polymerases (57, 58). A human cell has about 105 DNA lesions per day, the majority of which are able to block DNA polymerases (59, 60). Furthermore, genomic DNA harbors a great number of sites capable of forming DNA secondary structures which can act as native replication barriers. For example, human genomic DNA contains ∼700,000 potential G4 structures, plus a great number of potential hairpin and triplex forming sequences (61–63). Secondary DNA structures can form particularly on the DNA template for lagging strand synthesis because a single-stranded region of ∼150 nt or more is available on the lagging strand template due to the time gap between the unwinding of the double-stranded DNA (dsDNA) template and the subsequent initiation of lagging strand DNA synthesis. Thus, the stalling of DNA polymerases is likely a frequent event during DNA replication.
In this study, through a large scale of genetic screening, Cdc45, a subunit of the CMG complex, was identified as a potential checkpoint target: the cdc45R319I mutated cells had an almost normal cell growth cycle when compared to cdc45+ cells, but the mutation could dramatically reduce the sensitivity of cds1Chk2△ cells to 2.5 mM HU (Fig. 1A). Subsequent work demonstrated that, under conditions of fork stalling, Cds1Chk2 concurrently phosphorylated Cdc45 on S275, S322, and S397 (Fig. 1 B–G). The phospho-mimic and nonphosphorylation mutations, cdc45-3D and cdc45-3A, respectively, both dramatically reduced the sensitivity of cds1Chk2△ cells to 2.5 mM HU (∼600- to 3,000-fold; Fig. 2C) and to 1.0 mM stavudine (∼25-fold; Fig. 2E). The profound reduction in the sensitivity of cds1Chk2△ cells to HU or stavudine strongly suggests that Cdc45/CMG is a crucial checkpoint target in the checkpoint-mediated stabilization of stalled forks. The response to stavudine, the incorporation of which completely blocks replication, strongly indicates that cdc45-3D and cdc45-3A mutations increase the stability of stalled forks rather than simply reducing the rate of dNTPs consumption during HU-induced fork stalling. Consistent with this, we show that interorigin distances are unchanged in the early S phase in cdc45-3D and cdc45-3A cultures when compared to cdc45+, demonstrating there is no systemic initiation defect in these cells.
We next demonstrated that Cds1Chk2-mediated phosphorylation of Cdc45 on S275, S322, and S397 dramatically reduced CMG helicase activity (Fig. 5 D and H) and verified that Cds1Chk2 checkpoint kinase-dependent slowing of replication in the presence of HU occurred in S. pombe (Fig. 4 E, Middle). The stress-induced slowing of replication fork progression is consistent with the slowing of replication under replication stress that was observed by Paulovich and Hartwell (36). The rate of replication elongation, or fork speed, depends largely on replicative helicase activity and on the degree of chromatin compaction: replication fork speed in eukaryotic cells is ∼20 nt/s, but the nt incorporation rates by DNA polymerase δ and ε on naked DNA are ∼400 nt/s (64). In a previous study, we showed that the chromatin surrounding stalled replication forks becomes more compact and that this compacted chromatin contributes to preventing the CMG helicase from moving away from DNA polymerases/replication forks (Chromsfork control) (44). This current work answers the following question: Does the intra-S phase checkpoint target any other factors to slow down DNA replication under replication stress? While a number of replication components have been reported to be phosphorylated by the checkpoint in response to fork stalling or DNA damage, including RPA, Dna2 (17, 65–67), and MCM subunits (68–70), none of these modifications have been linked directly to the regulation of replication fork speed. Our identification of the regulation of the CMG helicase by Cdc45 phosphorylation demonstrates that cells use at least two mechanisms to prevent the CMG helicase from delinking from DNA polymerases. One is Chromsfork-mediated chromatin compaction around stalled forks and the other is the checkpoint-mediated reduction of CMG helicase activity.
Unusually for the analysis of phosphorylation site mutants, both the cdc45-3A and cdc45-3D alleles mimic the checkpoint regulation by stabilizing stalling replication forks. A simple explanation for this is that Cds1Chk2-mediated phosphorylation of Cdc45 on S275, S322, and S397, and cdc45-3D and cdc45-3A mutations all reduce the CMG helicase activity (Fig. 5). Why does the cdc45-3A phospho-null mutation, like the cdc45-3D phospho-mimic, reduce the CMG helicase activity? We propose that the reason pertains to a unique biochemical property of CMG helicase. CMG is composed of Cdc45, MCM, and GINS. MCM is the catalytic component, but it alone does not have detectable ATPase and helicase activity (71). Once associated with Cdc45 and GINS, MCM, or actually CMG, becomes a robust ATPase and helicase (72–76). This demonstrates that robust CMG helicase activity requires proper interactions among Cdc45, MCM, and GINS. Based on recently resolved structures of human Cdc45 and GINS and Saccharomyces cerevisiae CMG at a resolution of 3.7 to 4.8 Å (77–79), the S275, S322, and S397 residues of Cdc45 lie in the CID domain that is responsible for interacting with Mcm2, Mcm5, Psf1, and Psf2. In support of a core role for these serine residues in coordinating Cdc45-CMG interactions, the mutants cdc45-3C, cdc45-3K, cdc45-3V, cdc45-3N, and cdc45-3G also caused otherwise wt cells to become sensitive to HU and significantly rescued the HU sensitivity of cds1Chk2△ cells (SI Appendix, Fig. S1 H and I). Importantly, it has also been observed in other proteins that the phenotype of the phosphor-mimic mutation (serine or threonine to aspartate) is recaptured by the mutation of serine or threonine to alanine (80, 81).
The importance of the S275, S322, and S397 residues for CMG helicase activity is further evidenced by our observation that a cdc45-3T allele (three serines changed to threonines) caused slightly increased sensitivity to 5.0 or 7.5 mM HU when compared to cdc45+ and moderately rescued the sensitivity of the cds1Chk2△ cells to 1.5 mM HU (SI Appendix, Figs. S1 H and I and S5D). This implies that the S275, S322, and S397 residues are critical for Cdc45 to interact properly with MCM and GINS for a normal level of CMG helicase activity. This could explain why the phosphorylation of these three sites, or mutation to aspartic acid (cdc45-3D), alanine (cdc45-3A), and glutamic acid (cdc45-3E), all significantly reduced the helicase activity of CMG (Fig. 5 B and D) and rescued the sensitivity of cds1Chk2△ cells to HU (Fig. 2 C and D; note, we were unable to examine cdc45-3E in serial dilution assays because the cdc45-3E cells grew very poorly [SI Appendix, Figs. S1 H and I and S4 A–C]). Taken together, our data indicate that the intra-S phase checkpoint regulates Cdc45 to decrease the helicase activity of CMG in response to fork stalling. The S275, S322, and S397 sites within the CID domain of Cdc45 provide an ideal mechanism to regulate CMG helicase activity because even a very minor change in these three serines, such as a mutation to threonine, affects the CMG helicase activity.
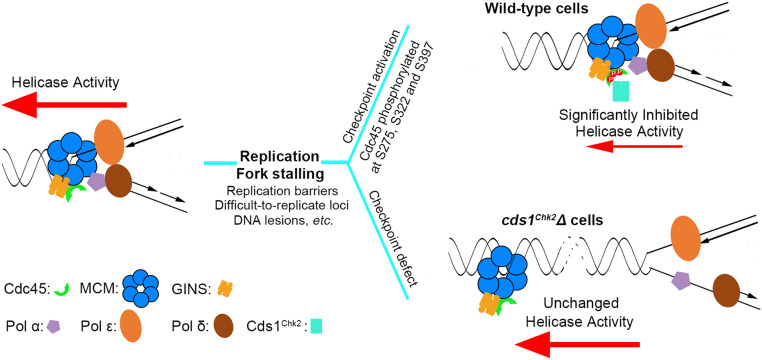
Why does the stabilization of stalled replication forks require the reduction of replicative helicase activity? The answer to this question should account for the fundamental biochemical reactions occurring at forks. Normally, the helicase-mediated unwinding of template dsDNA must be strictly coordinated with the synthesis of the leading and lagging strands. This coordination is achieved through protein interactions between the replicative helicase and DNA polymerases. In Escherichia coli, this is mediated by the τ subunit of the replisome and in eukaryotes by the Ctf4 and Mrc1 proteins (26, 82, 83). However, when (as stated above) DNA polymerases are blocked during DNA replication, the replicative helicase, powered by ATP hydrolysis, can move forward inappropriately and delink from the DNA polymerases. This would, in effect, collapse forks: once CMG moves away from a fork without being closely followed by DNA polymerization, the two unwound, and complementary DNA strands behind the helicase are vulnerable to nucleases and also can potentially reanneal, allowing nucleosomes to reassemble on the reannealed dsDNA, making the separation of replicative helicase and DNA polymerases unreversible (Fig. 7). Thus, when DNA polymerases are blocked, the intra-S phase checkpoint Cds1Chk2 kinase concurrently phosphorylates Cdc45 on the S275, S322, and S397 sites and dramatically reduces the CMG helicase activity. This, along with other responses such as Chromsfork, prevents the CMG helicase delinking from DNA polymerases and contributes to preserving the integrity of replisomes.
Fig. 7.
Cds1Chk2 phosphorylates Cdc45 to inhibit CMG helicase activity, reduce fork speed, and prevent physical separation of the CMG helicase and DNA polymerases in stalling/stalled replication forks. This results in the stabilization of stalling replication forks. (Left) As replication forks traverse DNA, the polymerases act coordinately with the CMG helicase to synthesize the leading and lagging strands. When replication polymerases are impeded, a small amount of ssDNA is revealed, which activates the intra-S phase ATR-dependent checkpoint pathway (Top, Right). Activated Cds1Chk2 phosphorylates the Cdc45 subunit of the CMG replicative helicase complex on S275, S322, and S397. This results in a severe reduction of CMG helicase activity and prevents the physical uncoupling of strand separation and DNA polymerization, thus stabilizing the stalling replication fork. (Bottom, Right) In checkpoint-deficient cells, CMG helicase activity is not inhibited, which results in DNA unwinding proceeding faster than DNA polymerization, the exposure of excessive ssDNA, and fork collapse.
Replication stress not only affects genomic stability but also threatens cell survival. Thus, several distinct but closely related mechanisms of the intra-S phase checkpoint regulation have been developed to cope with replication stress. Besides its critical role in stabilizing stalling replication forks, the intra-S phase checkpoint also regulates to block mitosis (84, 85) in order to avoid catastrophe caused by cell division when DNA replication is incomplete. Furthermore, it has been well documented that the intra-S phase checkpoint regulates to inhibit late origin firing when cells suffer replication stress (31–34, 86). The inhibition of late origin firing reduces the number of replication forks encountering stress, thus alleviating the replication stress of cells; preventing the exhaustion of RPA (and possibly some other replication factors as well) is another reason for checkpoint regulation to inhibit late origin firing, thus prohibiting replication catastrophe (34).
Materials and Methods
The information of strains, plasmids, antibody generation or purchased antibodies, the preparation of cell or chromatin extracts, DNA combing assay, the isolation of replication forks, the isolation of the replicative helicase CMG complex from chromatin, expression and purification of S. pombe Cds1 and Cdc45, EdU incorporation and detection, and assay of in vitro CMG helicase activity are in SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the D.K. laboratory for discussions and support, Xiao-chen Li and Hong-xia Lv for help with fluorescent assays, and Li-lin Du for genetic advice. This work was supported by grants from the National Natural Science Foundation of China (Nos. 31230021 and 31170739), the Ministry of Science and Technology of China (Nos. 2013CB911000 and 2010CB912201), the Peking-Tsinghua Center for Life Sciences, and the National Key Laboratory of Protein and Plant Gene Research. We thank the National Center for Protein Sciences (Beijing) at Peking University for assistance with FACS and microscopic assays. A.M.C. acknowledges Medical Research Council (United Kingdom) Grant G1100074.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.-S.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019183118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Torres J. Z., Schnakenberg S. L., Zakian V. A., Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: Viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 24, 3198–3212 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirkin E. V., Mirkin S. M., Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 71, 13–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuno K., Miyabe I., Schalbetter S. A., Carr A. M., Murray J. M., Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493, 246–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilera A., García-Muse T., Causes of genome instability. Annu. Rev. Genet. 47, 1–32 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Cha R. S., Kleckner N., ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297, 602–606 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Paeschke K., Capra J. A., Zakian V. A., DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145, 678–691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minca E. C., Kowalski D., Replication fork stalling by bulky DNA damage: Localization at active origins and checkpoint modulation. Nucleic Acids Res. 39, 2610–2623 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maga G., van Loon B., Crespan E., Villani G., Hübscher U., The block of DNA polymerase delta strand displacement activity by an abasic site can be rescued by the concerted action of DNA polymerase beta and Flap endonuclease 1. J. Biol. Chem. 284, 14267–14275 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorgoulis V. G., et al., Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Bartkova J., et al., DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Macheret M., Halazonetis T. D., Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 555, 112–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aird K. M., Zhang R., Nucleotide metabolism, oncogene-induced senescence and cancer. Cancer Lett. 356, 204–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert S., Watson A., Sheedy D. M., Martin B., Carr A. M., Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121, 689–702 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Zeman M. K., Cimprich K. A., Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasetti C., Vogelstein B., Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasetti C., Li L., Vogelstein B., Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J., et al., The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 149, 1221–1232 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Tercero J. A., Diffley J. F., Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412, 553–557 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Sogo J. M., Lopes M., Foiani M., Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Cobb J. A., Bjergbaek L., Shimada K., Frei C., Gasser S. M., DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22, 4325–4336 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucca C., et al., Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23, 1206–1213 (2004). [DOI] [PubMed] [Google Scholar]
- 22.De Piccoli G., et al., Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol. Cell 45, 696–704 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Katou Y., et al., S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Noguchi E., Noguchi C., McDonald W. H., Yates J. R. III, Russell P., Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 24, 8342–8355 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naylor M. L., Li J. M., Osborn A. J., Elledge S. J., Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc. Natl. Acad. Sci. U.S.A. 106, 12765–12770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lou H., et al., Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell 32, 106–117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka K., Russell P., Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3, 966–972 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Kai M., Boddy M. N., Russell P., Wang T. S., Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 19, 919–932 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyabe I., Morishita T., Shinagawa H., Carr A. M., Schizosaccharomyces pombe Cds1Chk2 regulates homologous recombination at stalled replication forks through the phosphorylation of recombination protein Rad60. J. Cell Sci. 122, 3638–3643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couch F. B., et al., ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27, 1610–1623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santocanale C., Diffley J. F., A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395, 615–618 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Zegerman P., Diffley J. F., Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467, 474–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Mosqueda J., et al., Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467, 479–483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toledo L. I., et al., ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Shirahige K., et al., Regulation of DNA-replication origins during cell-cycle progression. Nature 395, 618–621 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Paulovich A. G., Hartwell L. H., A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82, 841–847 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Sabatinos S. A., Green M. D., Forsburg S. L., Continued DNA synthesis in replication checkpoint mutants leads to fork collapse. Mol. Cell. Biol. 32, 4986–4997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enoch T., Carr A. M., Nurse P., Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 6, 2035–2046 (1992). [DOI] [PubMed] [Google Scholar]
- 39.Lambert S., Froget B., Carr A. M., Arrested replication fork processing: Interplay between checkpoints and recombination. DNA Repair (Amst.) 6, 1042–1061 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Carr A. M., Paek A. L., Weinert T., DNA replication: Failures and inverted fusions. Semin. Cell Dev. Biol. 22, 866–874 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Branzei D., Foiani M., Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Smith J., Tho L. M., Xu N., Gillespie D. A., The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108, 73–112 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Lindsay H. D., et al., S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12, 382–395 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng G., et al., Replication fork stalling elicits chromatin compaction for the stability of stalling replication forks. Proc. Natl. Acad. Sci. U.S.A. 116, 14563–14572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nitani N., Nakamura K., Nakagawa C., Masukata H., Nakagawa T., Regulation of DNA replication machinery by Mrc1 in fission yeast. Genetics 174, 155–165 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Neill T., et al., Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J. Biol. Chem. 277, 16102–16115 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Seo G. J., et al., Determination of substrate specificity and putative substrates of Chk2 kinase. Biochem. Biophys. Res. Commun. 304, 339–343 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Lea A. P., Faulds D., Stavudine: A review of its pharmacodynamic and pharmacokinetic properties and clinical potential in HIV infection. Drugs 51, 846–864 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Jackson D. A., Pombo A., Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong Y., et al., The level of origin firing inversely affects the rate of replication fork progression. J. Cell Biol. 201, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer D. R., Das S., Rhind N., Analysis of DNA replication in fission yeast by combing. Cold Spring Harb. Protoc. 2018, 199–207 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Heichinger C., Penkett C. J., Bähler J., Nurse P., Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 25, 5171–5179 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mickle K. L., et al., Checkpoint independence of most DNA replication origins in fission yeast. BMC Mol. Biol. 8, 112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lundin C., et al., Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33, 3799–3811 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunkel T. A., Erie D. A., Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kornberg A., DNA replication. J. Biol. Chem. 263, 1–4 (1988). [PubMed] [Google Scholar]
- 57.Tissier A., et al., Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase iota. EMBO J. 19, 5259–5266 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trakselis M. A., Cranford M. T., Chu A. M., Coordination and substitution of DNA polymerases in response to genomic obstacles. Chem. Res. Toxicol. 30, 1956–1971 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Saul R. L., Ames B. N., Background levels of DNA damage in the population. Basic Life Sci. 38, 529–535 (1986). [DOI] [PubMed] [Google Scholar]
- 60.Tubbs A., Nussenzweig A., Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krasilnikova M. M., Mirkin S. M., Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 24, 2286–2295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gellon L., et al., Mrc1 and Tof1 prevent fragility and instability at long CAG repeats by their fork stabilizing function. Nucleic Acids Res. 47, 794–805 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chambers V. S., et al., High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 33, 877–881 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Mondol T., Stodola J. L., Galletto R., Burgers P. M., PCNA accelerates the nucleotide incorporation rate by DNA polymerase δ. Nucleic Acids Res. 47, 1977–1986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Recolin B., Maiorano D., Implication of RPA32 phosphorylation in S-phase checkpoint signalling at replication forks stalled with aphidicolin in Xenopus egg extracts. Biochem. Biophys. Res. Commun. 427, 785–789 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Stokes M. P., et al., Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elia A. E., et al., RFWD3-dependent ubiquitination of RPA regulates repair at stalled replication forks. Mol. Cell 60, 280–293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cortez D., Glick G., Elledge S. J., Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 101, 10078–10083 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoo H. Y., Shevchenko A., Shevchenko A., Dunphy W. G., Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J. Biol. Chem. 279, 53353–53364 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Yamada M., et al., ATR-Chk1-APC/CCdh1-dependent stabilization of Cdc7-ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress. Genes Dev. 27, 2459–2472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishimi Y., A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 272, 24508–24513 (1997). [DOI] [PubMed] [Google Scholar]
- 72.Costa A., et al., The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18, 471–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kubota Y., et al., A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 17, 1141–1152 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gambus A., et al., GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C., Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21, 581–587 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Moyer S. E., Lewis P. W., Botchan M. R., Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simon A. C., Sannino V., Costanzo V., Pellegrini L., Structure of human Cdc45 and implications for CMG helicase function. Nat. Commun. 7, 11638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang Y. P., Wang G., Bermudez V., Hurwitz J., Chen X. S., Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc. Natl. Acad. Sci. U.S.A. 104, 12685–12690 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan Z., et al., Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 23, 217–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dai H., et al., Contribution of Bcl-2 phosphorylation to Bak binding and drug resistance. Cancer Res. 73, 6998–7008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McClure A. W., Diffley J. F. X., Rad53 checkpoint kinase regulation of DNA replication fork rate via Mrc1 phosphorylation. bioRxiv [Preprint] (2021). 10.1101/2021.04.09.439171 (Accessed 13 April 2021). [DOI] [PMC free article] [PubMed]
- 82.McHenry C. S., DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 80, 403–436 (2011). [DOI] [PubMed] [Google Scholar]
- 83.Simon A. C., et al., A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510, 293–297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeng Y., et al., Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature 395, 507–510 (1998). [DOI] [PubMed] [Google Scholar]
- 85.Boddy M. N., Furnari B., Mondesert O., Russell P., Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280, 909–912 (1998). [DOI] [PubMed] [Google Scholar]
- 86.Dungrawala H., et al., The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.