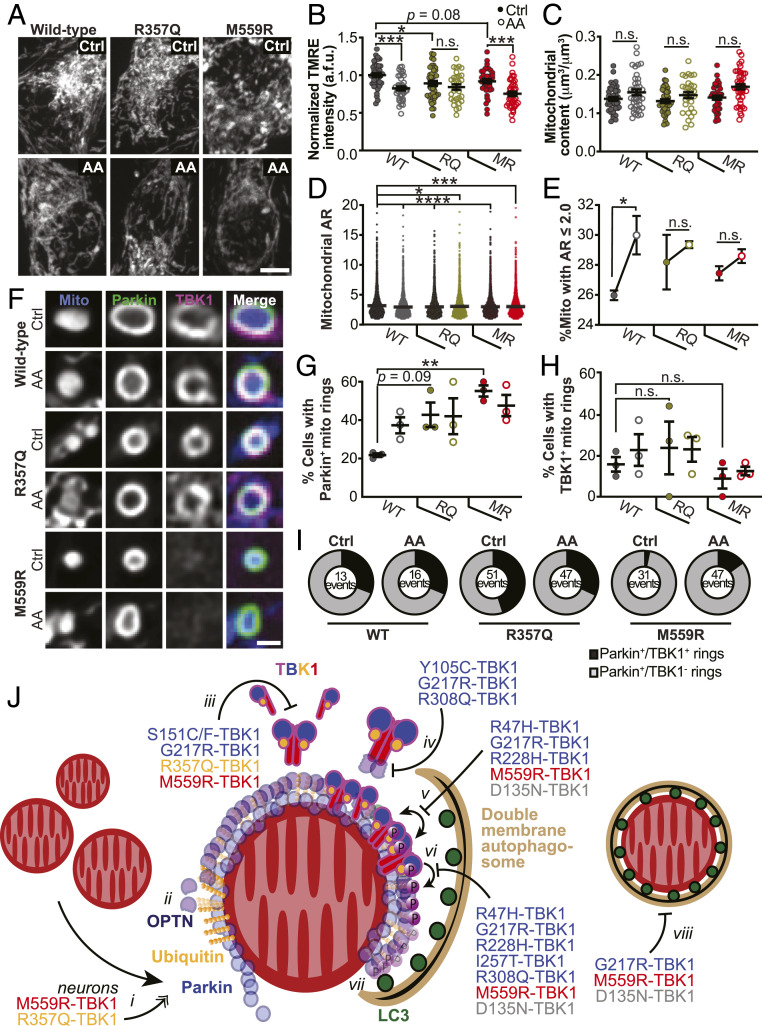
Fig. 8.
Expression of ALS-associated TBK1 mutants alters mitochondrial network health and sensitivity to oxidative stress, and a model for the deleterious effects of TBK1 mutations in mitophagy. (A and B) Representative images (A) and quantification (B) of TMRE fluorescence intensity. Control basal conditions, Ctrl. Mean SEM; n = 30 to 42 neurons from three to four biological replicates; 7 days in vitro (DIV). Not significant, n.s.; *P ≤ 0.05, ***P < 0.001 by one-way ANOVA with Sidak’s multiple comparisons test. (Scale bar, 5 μm.) (C) Quantification of mitochondrial content by volume. Mean SEM; n = 30 to 42 neurons from three to four biological replicates; 7 DIV. Not significant, n.s., by Kruskal–Wallis ANOVA with Dunn’s multiple comparisons test. (D) Quantification of mitochondrial AR for all mitochondria observed. Mean SEM; n = 30 to 42 neurons from three to four biological replicates; 7 DIV. *P ≤ 0.05, ***P < 0.001, ****P < 0.0001 by one-way ANOVA with Dunnett’s multiple comparisons test. (E) Percent of mitochondria with a mitochondrial AR 2. Mean SEM; n = 30 to 42 neurons from three to four biological replicates; 7 DIV. Not significant, n.s.; *P ≤ 0.05 by one-way ANOVA with Sidak’s multiple comparisons test. (F) Representative images of Parkin-positive mitochondria with examples that are TBK1 positive (WT and R357Q) and TBK1 negative (M559R). (Scale bar, 1 μm.) (G and H) Quantification of the percent of neurons with Parkin-positive (G) or TBK1-positive (H) mitochondria rings. Mean SEM; n = 25 to 32 neurons from three biological replicates; 7 DIV. Not significant, n.s.; **P < 0.01 by one-way ANOVA with Sidak’s multiple comparisons test. (I) Quantification of the number of Parkin-positive mitochondria (total) that are TBK1 positive (black sector) or TBK1 negative (gray sector). n = 25 to 32 neurons from three biological replicates; total number of events are shown; 7 DIV. (J) Model for TBK1 involvement in mitophagy and effects of mutants. i) Upon mitochondrial depolarization, Parkin (blue circles) is recruited to the OMM and ubiquitinates (gold spheres) outer membrane proteins. In neurons, expression of R357Q- or M559R-TBK1 induces more rounded, Parkin-positive mitochondria. ii) Ubiquitin chains recruit OPTN (purple circles), which interact with ubiquitin via their UBAN domains. TBK1 is not required for this interaction. iii) TBK1 (multicolored cartoon) monomers constitutively dimerize along their scaffolding dimerization domains. Five mutations disrupt this dimerization, including R357Q-TBK1 and M559R-TBK1, which have completely disrupted dimerization. iv) TBK1 associates with OPTN at its CTD, thus TBK1 may be corecruited with OPTN to the mitophagy site. Three ALS-linked mutations in TBK1 exhibit disrupted OPTN association, yet Y105C-TBK1 and R308Q-TBK1 can still be recruited to the damaged mitochondria; thus TBK1 can also be independently recruited. v) Formation of TBK1 multimers at the mitochondria surface promotes TBK1 transautophosphorylation, by which TBK1 is activated upon phosphorylation at S172 (purple circles with “P”). Four ALS-linked TBK1 mutants and the engineered D135N-TBK1 have diminished or abolished activation. vi) Activated TBK1 phosphorylates the mitophagy receptor OPTN at S177. Activated TBK1 may also promote autophagosomal membrane expansion (tan crescent). vii) Phosphorylated OPTN is necessary to recruit the LC3-coated (dark green circles) autophagosome. viii) The double membrane autophagosome completely engulfs a damaged mitochondria.