Abstract
Objective
Among the stakeholders of COVID-19 research, clinicians particularly experience difficulty keeping up with the deluge of SARS-CoV-2 literature while performing their much needed clinical duties. By revealing major topics, this study proposes a text-mining approach as an alternative to navigating large volumes of COVID-19 literature.
Materials and methods
We obtained 85,268 references from the NIH COVID-19 Portfolio as of November 21. After the exclusion based on inadequate abstracts, 65,262 articles remained in the final corpus. We utilized natural language processing to curate and generate the term list. We applied topic modeling analyses and multiple correspondence analyses to reveal the major topics and the associations among topics, journal countries, and publication sources.
Results
In our text mining analyses of NIH's COVID-19 Portfolio, we discovered two sets of eleven major research topics by analyzing abstracts and titles of the articles separately. The eleven major areas of COVID-19 research based on abstracts included the following topics: 1) Public Health, 2) Patient Care & Outcomes, 3) Epidemiologic Modeling, 4) Diagnosis and Complications, 5) Mechanism of Disease, 6) Health System Response, 7) Pandemic Control, 8) Protection/Prevention, 9) Mental/Behavioral Health, 10) Detection/Testing, 11) Treatment Options. Further analyses revealed that five (2,3,4,5, and 9) of the eleven abstract-based topics showed a significant correlation (ranked from moderate to weak) with title-based topics.
Conclusion
By offering up the more dynamic, scalable, and responsive categorization of published literature, our study provides valuable insights to the stakeholders of COVID-19 research, particularly clinicians.
Keywords: COVID-19, SARS COV-2, Text mining, Natural language processing, Topic modeling
Graphical abstract

Introduction
Since its emergence in Wuhan, China, in December 2019, COVID-19 has spread across the world and generated 28 million confirmed cases and a death toll of 2.1 million as of January 21, 2021 [1]. In response to this public health emergency, numerous open-access research data, and computational resources have been established by the National Institutes of Health (NIH), Centers for Disease Control and Prevention (CDC), public consortia, and some private entities [[2], [3], [4]]. Consequently, an unprecedented rate of growth in the literature related to COVID-19 and associated implications has emerged. To identify relevant information from the abundance of COVID-19 articles, the White House Office of Science and Technology Policy initiated a call to action to the artificial intelligence (AI) community [3].
Thus far, numerous AI techniques have been applied to population screening, imaging data acquisition, diagnosis, and medical support for COVID-19 [[5], [6], [7]]. Bibliometric analysis of research results has been conducted to investigate the current status of published COVID-19 research [[8], [9], [10], [11]]. Colavizza et al. conducted a scientometric analysis to map scientific literature [12]. Other studies incorporated text-mining techniques such as Lexical Link analysis [13], Named Entity Recognition [14], topic modeling analysis [15], and Latent Dirichlet Allocation [16,17] to identify and visualize the topics and patterns of COVID-19 literature. However, these studies either had a larger focus of all coronavirus infection types, including severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV) [15,16] or, in the case of some bibliometric studies, included very limited numbers of studies−ranging from 92 to 923− with a COVID-19 focus. Work to date has not yet produced a set of topics around which the literature can be organized and searched. In this study, we examine tens of thousands of articles to develop such a topic set through the use of title-based and abstract-based models and the examination of the relationships between them.
Materials and methods
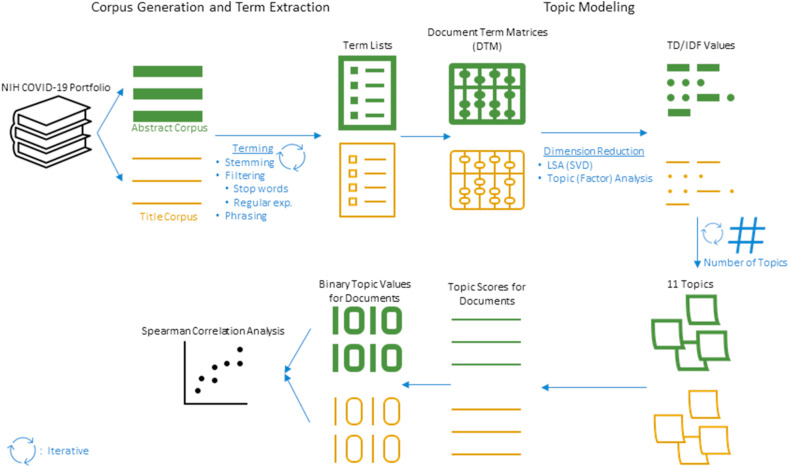
This study preprocesses a corpus of articles to extract terms and then apply a series of content analysis methods [[18], [19], [20]]. Our study comprised two parts. In the first part, we identified a resource for COVID-19 literature and developed our initial corpus for curation and term extraction. In the second part, we utilized several dimension reduction techniques including topic modeling and multiple correspondence analysis to reveal some distinct patterns in the text data and the associations among these patterns. Given that the study sample included publicly available research abstracts and titles, this study did not require institutional review board review. Human subjects were not used for this study. Fig. 1 illustrates the steps involved in this study, starting from the establishment of corpora for abstracts and titles separately from NIH COVID-19 Portfolio. The green and yellow colors indicate processes focusing on abstracts and titles, respectively. This initial step is followed by the generation of term lists using various NLP techniques and the establishment of DTMs. The next step involves the use of dimension reduction techniques and the determination of numbers of topics for abstracts and titles. The final step involves processes to generate topic scores and associated binary values, then merging results for abstract-based and title-based topics to examine the Spearman correlation between them.
Fig. 1.
Topic Modeling Steps. The green and yellow color indicate processes for abstracts and titles, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Corpus generation and term extraction
We downloaded all references from the NIH COVID-19 Portfolio, an expert-curated source for publications and preprints for COVID-19 or the novel coronavirus SARS-CoV-2 [21], on July 25. We, then updated the set of references on November 10th and 21st before we performed the final analyses. We excluded references in which abstracts were brief (less than 24 words) or absent.
To curate and generate the term list, we used natural language processing (NLP) methods such as stop words and regular expressions−exclusion of unimportant words and patterns that are irrelevant to the study; phrasing − recognizing phrases up to six words (6-g) such as severe acute respiratory syndrome coronavirus-2 (SARS-COV-2); and stemmitazion/lemmatization − reducing terms into their simplest form (i.e., roots) [19]. We used JMP 14 Pro Text Explorer for data curation and analyses [22] due to its ability to process large corpora without requiring high computational power like a super computer and its user-friendly interface that do not require prior programming experience. We produced two document term matrices (DTMs), one for the abstracts and one for the titles.
Topic modeling
We utilized several dimension reduction techniques on the DTMs to discover patterns − major topics − by analyzing the abstracts and titles separately. First, we used latent semantic analysis (LSA), which uses singular value decomposition (SVD), a matrix decomposition process to reduce a matrix into its constituent parts, to reveal principle components of the matrices [23,24]. Then, we used topic analyses (TA), a technique similar to factor analysis, to determine the optimum number of topics between 8 and 16, by examining the terms with higher TF-IDF (term frequency-inverse document frequencies) weights. In LSA and TA, researchers determines the numbers of topics by running the analyses using pre-determined topic numbers and examining the resulted terms in each different topic models. The visual cues such as negative loaded terms in JMP Text Explorer allow the researcher to find the optimum number of topics. Negatively loaded term indicates that the particular term does not belong to the topic that it was included. To find a home for a negatively loaded term, researchers would increase the number of topics and examine the results. In our case, we examined the results starting from 8-topic to 16-topic models for abstracts. After determining 11 as the best number for abstract-based topics, we examined the title-based topics, which also exhibited similar results. Since we aimed to investigate the correlation between topics generated from abstracts and titles, we determined the title-based topics as 11 to achieve one-on-one matching. We preferred TF-IDF as the weighting factor over regular frequency due to its normalization function, highlighting the unbiased importance of a particular term while considering the entire corpus [18]. Topics are ranked from 1 to 11 according to the amount of variation explained by a particular topic in the respective data set [25]. The topics were named by the study authors, including four medical doctors, by using the high-frequency terms for each topic.
After determining an 11-topic model for abstracts and another for titles, we assigned each document (publication) to an abstract-based topic and to a title-based topic based on the document's highest topic score (i.e., component score). Topic scores are a vector of orthogonal values for each document that was generated by extracting and summarizing the patterns from the original DTM [25]. Documents that achieve a higher score in a topic would exhibit higher association with that particular topic [22]. Based on the highest topic score for each document, we generated binary values for each of the eleven topics for abstracts and titles. In each of the abstract and title analysis file, we ended up with a binary value for each topic that describe a document's belonging to that topic. We then merged the two analysis files using unique document ID numbers. We examined the correlation (Spearman) between 11 abstract- and 11 title-based topics using the binary variables. We generated word clouds for top loading terms for each topic, and examined the content of the word clouds to assess the semantic compatibility between abstract-based and title-based topics that exhibited some level of correlation. We also examined the relationship between abstract-based 11 topics and their sources and journal countries by using multiple correspondence analysis; a technique stems from the principal component analysis designed for categorical variables [26]. Multiple correspondence analysis shows the associations between categorical variables. Finally, we developed a table that included the five most representative articles (based on highest-ranking of topic scores) for each of the eleven abstract-based topics to provide further insights.
Results
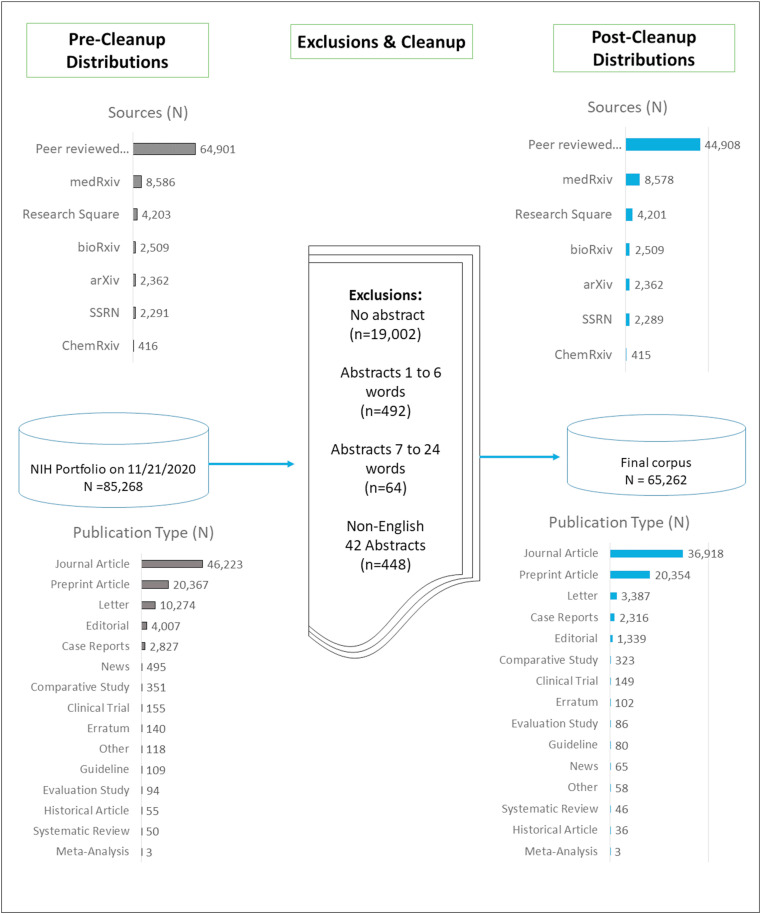
We were able to obtain 85,268 references from the NIH COVID-19 Portfolio [21] as of November 21. After the exclusion based on inadequate abstracts, 65,262 articles remained in the final corpus. Given that one of the goals of the study was to compare topics generated from abstracts and titles separately, we excluded the references that did not include abstracts and abstracts with less than 24 words. We further excluded 42 non-english abstracts to reduce potential noise in the data. Our results can be reproduced by other researchers by following the exclusion criteria highlighted in Fig. 2 . The resulting abstract and title DTMs included 9,923 and 2,512 terms, respectively. Fig. 2 displays the corpus generation process and pre/post distributions of publications indicating their sources and types.
Fig. 2.
Corpus generation process, and pre/post distributions of references based on 1) sources, and 2) article types. Covid-19 Portfolio, a NIH initiative that combines articles from PubMed and preprints from arXiv, bioRxiv, ChemRxiv, and medRxiv, Research Square, and SSRN, was downloaded on 11/21/2020 from https://icite.od.nih.gov/covid19/search/.
Fig. 3, Fig. 4 illustrate the word cloud results for eleven topics based on the frequency of terms commonly used within the COVID-19 literature. The font size of terms is based on the TF/IDF scores, in which higher scores refer to larger font size. Each cluster in the word clouds exhibit the top 40 loading terms for the topic. Some terms in the clouds are displayed with a dot at the end to indicate they were stemmed -reduced into the root of the word. Topics were generated for both abstracts and titles (Fig. 3, Fig. 4, and Appendix A). Identified topics in abstracts are 1) Public Health, 2) Patient Care & Outcomes, 3) Epidemiologic Modeling, 4) Diagnosis and Complications, 5) Mechanism of Disease, 6) Health System Response, 7) Pandemic Control, 8) Protection/Prevention, 9) Mental/Behavioral Health, 10) Detection/Testing, 11) Treatment Options. Fig. 3 also displays the correlations between topics based on abstracts and titles (5 topics for each). The visual examination of the word clouds for the topics with moderate and weak correlations suggests the semantic compatibility between titles and abstracts of the publications. Due to their similarities, the corresponding -abstract and title based-topics were named identically.
Fig. 3.
Moderate and weak correlated topics based on abstracts and titles. Word clouds exhibiting low weak to moderate correlations between topics based on: 1) abstracts (A), and 2) titles (T). The topics based on abstract and the corresponding title-based ones received the same name due to their similarity as evidenced by the similar terms exhibited in the word clouds. The size of font of a particular term indicates the term frequency inverse document frequency (TF-IDF) values. Different color spectrums were used for abstract-based topics and title-based topics. Therefore, similar colors within abstract-based word clouds indicates similar TF-IDF values (blue color being the indicative of high TF IDF value). A: Abstract, T: Title; Top: Topic; Top. A 9 & Top. T 5: Mental/Behavioral Health, Topic A 2 & Topic T 9: Patient Care and Outcomes, Topic A 3 & Topic T 8: Epidemiologic Modeling, Topic A 5 & Topic T 2: Mechanism of Disease, Topic A 4 & Topic T 11: Diagnosis and Complications. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
* Abstract-based topics exhibiting very small or no correlation to the title-based topics. Top. A 1: Public Health, Top. A 6: Health System Response, Top. A 7: Pandemic Control, Top. A 8: Protection/Prevention, Top. A 10: Detection/Testing, Top. A 11: Treatment Options. * The word clouds for the title-based topics exhibiting very small or no correlation to the abstract-based topics are provided in Appendix A.
Fig. 4displays the remaining six topics. These abstract-based topics exhibited very small or no correlations to the title-based topics. The remaining title-based topics are demonstrated in Appendix A.
The five references that achieved the highest topic score (i.e., principal component score) for every 11 abstract-based topics are provided in Appendix B. Each set of five articles can be considered as the most representative of their corresponding topic due to their inclusion of a higher proportion of topic-specific terms. These representative articles can be used for additional insights into the identified topic. In the following paragraph, the description of the first topic is provided as an example to interpret the results, shown in Fig. 3, Fig. 4 and Appendix A & B.
Topic A 1. public health
This topic includes 5,448 abstracts and consists of 9% of the entire corpus of 65,262 articles (Fig. 4). In this topic, some of the most frequently used terms (with the highest TF-IDF scores) are health, country, care, community, public, access, pandemic, and social (Fig. 4). The most representative five articles of topic A 1 mainly focus on public health, social, and educational implications of COVID-19 with different focuses such as “public health law and science” [27], “public health framework for COVID-19 business liability” [28], “COVID-19 Response and Education in South Africa” [29,30], and “challenges of scientific dissemination” [31] (Appendix B).
Fig. 5, the map of COVID-19 research, displays the results of the multiple correspondence analyses. All four maps show the association between abstract-based topic and another categorical variable. In all four maps the red and blue dots indicate the proportional size of the variable in terms of numbers of publication. In all maps, we inserted red squares around some stronger associations to highlight them and to draw the attention of the readers. The first map (a) shows the relationship between abstract-based topics and title-based topics. The highlighted associations with red squares in the first map (a) overlap with our Spearman correlation results in Fig. 3. The second map (b) illustrates the relationship between abstract-based topics and publication types. The third map (c) illustrates the relationship between abstract-based topics and the sources of the articles. Lastly, the fourth map (d) shows the relationship between the abstract-based topics and journal countries, which were available for only 44,876 abstracts. To improve visualization, we limited journal countries to the top 15 (n = 41,842, see Appendix C) and combined the remaining 3,034 articles from various countries into the “other” category.
Fig. 5.
Multiple correspondence analysis results exhibiting the associations among abstract-based topics and a) title-based topics, b) publication types, c) sources, d) journal countries*. The close proximity between particular topic and country/source indicates strong association between them. The size of the points indicates the proportional marker size, size of the points being proportional to the count of observations corresponding to each point. A: Abstract, T:Title; TopicA 1: Public Health, TopicA 2: Patient Care and Outcomes, TopicA 3: Epidemiologic Modeling, TopicA 4: Diagnosis and Complications, TopicA 5: Mechanism of Disease, TopicA 6: Health System Response, TopicA 7: Pandemic Control, TopicA 8: Protection/Prevention, TopicA 9: Mental/Behavioral Health, Topic A 10: Detection/Testing, TopicA 11: Treatment options. TopicT 1: Vaccine Development, TopicT 2: Mechanism of Disease, TopicT 3: Disease Management, TopicT 4: Treatment Protocols, TopicT 5: Mental/Behavioral Health, TopicT 6: Detection/Testing, TopicT 7: Diagnostic, TopicT 8: Epidemiologic Modeling, TopicT 9: Patient Care and Outcomes, TopicT 10: Health System Response, TopicT 11: Diagnosis and Complications. *For better visualization we included top 15 countries out of 58 journal countries in the analyses (See Appendix C).
Interpreting the results in Fig. 5 should be performed in combination with Fig. 1, Fig. 2, Fig. 3 and Appendix B. For example, Topic A 5, Mechanism of Disease, includes 5,218 articles and exhibits correlation with title-based topic 2 (Top. T 2) as illustrated both in Figs. 3 and 5 (a). Moreover, Topic A 5 exhibits stronger associations with journal articles and clinical trials as illustrated in Fig. 5 (b). To put this finding into the context, among all publication types, the journal article category in the COVID-19 portfolio has the highest proportion with 36,916 articles (Fig. 2). The location of Topic 5 A in Fig. 5 (c) indicates that this topic draws most of its content from peer reviewed (PubMed) articles. Despite their smaller size, the two archived sources, bioRxiv and ChemRxiv, also contribute to the Mechanism of Disease (Topic A 5). There is also a stronger association between Topic A 5, Mechanism of Disease, and Switzerland as illustrated in Fig. 5 (d). Lastly, Appendix B, the five most representative articles for each topic, includes three articles from Switzerland for Topic A 5 exploring issues related to mechanism of disease such as “immune dysregulation” [32], “non-invasive auricular vagus nerve simulation” [33], and “the role of zinc in the immunological pathways” [34]. Please see the maps to see the other relationships (Fig. 5).
Discussion
This study focuses on COVID-19, including 65,262 peer-reviewed and achieved studies through November 21, 2020, from NIH's COVID-19 portfolio. It provides a systematic assessment of organizing a voluminous literature accumulated in a matter of months using text mining techniques. Previous topic modeling studies on coronavirus-related research had an extensive focus, with studies ranging from as early as 1870 to April 23, 2020 [15,16]. However, having such a broad time range has the potential to divert the focus from COVID-19, the specific strain of the coronavirus infection that has resulted in a global pandemic in 2020. In contrast to the previous studies, this study focused on COVID-19, a specific strain of the coronavirus infection, identified eleven topics for COVID-19 research, contrasted the semantic compatibility of abstract versus title based topics, and mapped the COVID-19 research by showing the associations between those eleven topics, journal countries, and sources of publications.
For five of the 11 topics, we found moderate to weak correlations between topics that were generated using abstracts and titles. The visual examination of word clouds indicates high semantic compatibility between topics based on abstracts and titles suggests. This finding suggest that for a high-stakes, topic-modeling undertaking such as COVID-19 research, when all abstracts are not available, titles can be used to generate the major topical areas to inform the research community. To confirm this point, we further ran topic modeling analyses as sensitivity tests on the entire corpus of 85,268 titles before exclusions and found eleven topics that were very similar to the ones generated using our final corpus, which included 65,262 titles.
From clinicians' perspective, the finding of semantic compatibility of topics based on abstracts and titles is also valuable. Even though there have been many efforts to classify and organize COVID-19 literature [[35], [36], [37]], clinicians' time to explore COVID-19 literature is very limited due to the surge in COVID-19 cases. Even the current efforts to classify COVID-19 literature have grown into large numbers. These classification efforts tend to be very technical, making it difficult for clinicians to sift through while meeting the growing patient care demand. For example, the Kaggle site on the COVID-19 open research dataset challenge includes 17 tasks, encompassing 1,602 submissions [37]. The majority of these submissions are highly technical since they are geared towards developing new algorithms to achieve better searches. Therefore, to perform their day-to-day patient care using evidence-based medicine, clinicians still use default library search functions and review an article's title first before reviewing the abstract and full-text article. By showing the correlation between titles and abstracts, this study provides much insight to the clinicians about the reliability of using titles as the first step for developing a portfolio for evidence-based medicine.
Consistent use of terms in specific topics as visualized in word clouds (Fig. 3, Fig. 4) and the distinguishable themes for specific topics in the list of articles ranked from high to low relevancy for each topic (Appendix B) provides additional insights about topics and their consistency and integrity of our topic naming process. We found that for each topic in Appendix B the majority of articles explored a common, topic-specific issue. For example, all five articles in Patient Care and Outcomes (Topic A 2) focus on clinical care and patient outcomes, four of the five articles in Public Health (Topic A 1) focused on implications of COVID-19 on public health and education, and all five articles in Mental/Behavioral Health (Top A 9) explored issues such as stress, depression, seizures, and psychological needs.
There is some resemblance between our COVID-19 focused eleven topics, and the previous two studies of general coronavirus-focused topics. For instance, we were able to match most of the eight topics in a prior study [15] to our topics by examining the top-15 most frequent words. In contrast to the eight topics of this previous study [15], we found additional topics such as Mental/behavioral Health (Top. A 9) and Diagnosis and Complications (Top. A 4). Some of these additional topics, such as mental health, were also identified among fifty topics of a previous study [16]. Future studies may capture the evolution of COVID-19 research by comparing their results with our findings. There is the potential that some topics such as economic implications of the COVID-19 pandemic may become a major area of study as the pandemic evolves and such data are accumulated. Future studies may also explore monthly changes in topics during the pandemic and the potential lag time between practice and publication.
Stakeholders of COVID-19 research can draw valuable insights from the patterns of the research maps that we generated. Among eleven topics, Epidemiologic Modeling (Top. A 3) and Health System Response (Topic A 6) included the highest proportion (24%, and 13%), and Pandemic Control (Topic A 7) included the lowest proportion (4%) of articles. The strong association of peer reviewed sources and topics such as Protection/Prevention (Top. A 8), Treatment Options (Top. A 11), Diagnosis and Complications (Top. A 4), and Health System Response (Top. A 6) suggest that peer reviewed journals have a higher interest in these topics as opposed to topics such as Pandemic Control (Top. A7), Patient Care and Outcomes (Top. A 2), Epidemiologic Modeling (Top. A 3). On the other hand, the stronger association between archived sources and topics such as Pandemic Control (Top. A7), Patient Care and Outcomes (Top. A 2), Epidemiologic Modeling (Top. A 3) may also suggest a potential race among researchers to disseminate their findings.
The pattern of the two largest topics predominantly being published in two different venues suggests that more clinically focused health system response studies had a time advantage over Epidemiologic Modeling studies since the latter requires aggregation of data over time. The five representative articles (Appendix B) for Health System Response (Top. A 6) explored issues such as remote clinical skills, resident leadership, palliative care toolkit, management of the clinical and academic mission, and online trainee curriculum, none of which required extensive efforts for data collection. On the other hand, five representative articles (Appendix B) from Epidemiologic Models (Top. A 3) explored issues such as etiology of epidemic outbreaks, land-use change on livestock and implications on coronavirus outbreaks, wildlife supply chains for human consumption in Vietnam, and surveillance of bat coronaviruses in Korea. The authors of Epidemiologic Models seemed to overcome the time disadvantage by opting for achieved publishing, which takes less time compared to peer-reviewed publications. Additional evidence for earlier achieved publishing can also be found in Appendix B since the same study exploring the implication of wildlife supply chains for human consumption on transmission risk in Vietnam was initially published in bioRxiv, then in PLOS ONE [38]. We did not exclude preprint versions of same article to be able to see earlier achieved publishing pattern and check the validity of our text mining algorithm. Our text-mining algorithm placing the preprint and published version of the same article in the same topic also provides evidence of the integrity of our algorithm. When abstracts are very similar, or the same, the text mining algorithm places two documents into the same topics and calculates very similar topic scores for them.
Our findings on the relationship between topics and publication countries also provide additional insights. For example, the stronger association between Protection and Prevention (Top. A 8) with South Korea and the peer-reviewed sources is intuitive due to the well-publicized protection/prevention focus of the South Korean government. Moreover, the stronger association between topics such as Patient Care and Outcomes (Top. A 2), Health System Response (Top. A 6), Diagnosis and Complications (Top. A 4), and two countries, United States and Germany, suggest that publications from these countries focused more on clinical care issues during the pandemic.
Even though journal articles (n = 36,916) followed by preprint articles (20,354) represented the majority of the publication types in the entire corpus of 65,262, additional insights can be found while examining the associations between topics and publication types (Fig. 5 (b)). For example, the stronger association between preprint articles and Detection/Testing (Topic A 10) suggests the scientific race in disseminating viable study findings on detection/testing. This scientific dissemination race might be leading to a higher proportion of these publications being initially published as preprints. Another interesting insight was the strong association between Diagnosis and Complications (Topic A 4) and Case Reports. This phenomenon can also be observed in the five representative articles (Appendix B) on this topic. Two of the five articles were case reports exploring “a 44-year-old woman with chest pain, dyspnea, and shock” [39] and “Microthrombi and ST-Segment-Elevation Myocardial Infarction in COVID-19.” [40].
Limitations
First, we were not able to analyze author-provided keywords since they were not provided in NIH's COVID-19 portfolio, which instead included expert-curated columns for condition, chemicals & drugs, target, and devices. Given that abstracts contain a higher proportion of keywords compared to the other sections of articles [41], utilizing abstracts to generate topics was a methodologically sound approach. However, it would be valuable for future studies to explore the correlations among topics generated from abstracts, keywords, titles, and full-texts. Second, while generating topics and classifying documents into topics, there are some alternative methods such as manual human classification and Latent Dirichlet Allocation(LDA), a probabilistic model that identifies topics within the corpus and links the documents to those identified topics [42]. Even though there is a potential that these approaches could have achieved better accuracy, especially the manual human classification [43] one, we used LSA due to its dimension reduction focus and easier trainability, lesser time, and computational resource commitment. Despite its potential of higher accuracy, manual human topic generation and document classification was not a viable option since it would take a substantial amount of human resource and months to perform topic modeling for a corpus of 65,262 research articles. We did not have such human resources. There are also existing efforts led by the National Library of Medicine to curate and classify the literature using expert input [36]. However, we believe that computerized topic generation can be considered as a complement to manual human efforts since they can perform initial work by generating topics for experts to fine-tune and extract the most relevant information from those topics to enhance the application of evidence-based medicine. Another limitation pertains to the determination of the topic number. In traditional topic modeling approaches such as LSA, TA, and LDA, researchers determine the topic number through manual and iterative processes by running and examining the results starting from low to high topic models [44]. However, some of the recently developed topic modeling approaches such as Top2Vec, determine the optimum numbers of topics automatically [44]. However, these recent approaches require high computational power and tend to generate larger number of topics, which would fit better in studies focusing to reveal all potential topics, ranging from very minor to major ones. In future studies, we encourage the researchers to utilize methods other than LSA and compare their findings with our results.
Conclusion
Our study revealed major topics in COVID-19 literature by harnessing, synthesizing, and visualizing information from large volumes of research (N = 65,262) to inform the research community and evidence-based medicine. This is the first study that maps the relationships between abstract-based topics and four distinct categories, including 1) title-based topics, 2) publication types, 3) publication sources, and 4) publication countries. Stakeholders of COVID-19 research, particularly clinicians, would be able to draw valuable insights from the eleven major topics and the maps that exhibited associations between these topics, publication types, sources, and countries. Because of the rapid accumulation of COVID-19 manuscripts, text mining approaches are vital to increasing the likelihood that articles are found by those they have the most relevance to applying knowledge to optimize the global response.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ibmed.2021.100036.
Appendix A. Supplementary data
The following are the supplementary data to this article:
figs1.
References
- 1.Organization WH . 2020. Coronavirus disease (COVID-19) pandemic; pp. 6–25. [Google Scholar]
- 2.Health NIo . 2020, May 13. Open-access data and computational resources to address COVID-19. [Google Scholar]
- 3.House W. 2020, March 16. Call to action to the tech community on new machine readable COVID-19 dataset. [Google Scholar]
- 4.CORD-19 . 2020. COVID-19 open research dataset. [Google Scholar]
- 5.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev Biomed Eng. 2020:1. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 6.Vaishya R., Javaid M., Khan I.H., Haleem A. Artificial Intelligence (AI) applications for COVID-19 pandemic. Diabet Metab Syndr. 2020;14:337–339. doi: 10.1016/j.dsx.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hossain M.M. 2020. Current status of global research on novel coronavirus disease (Covid-19): a bibliometric analysis and knowledge mapping. Hossain MM Current status of global research on novel coronavirus disease (COVID-19): a bibliometric analysis and knowledge mapping. [version 1. [Google Scholar]
- 9.Dehghanbanadaki H., Seif F., Vahidi Y., Razi F., Hashemi E., Khoshmirsafa M., et al. Bibliometric analysis of global scientific research on Coronavirus (COVID-19) Med J Islam Repub Iran. 2020;34:354–362. doi: 10.34171/mjiri.34.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonilla-Aldana D.K., Quintero-Rada K., Montoya-Posada J.P., Ramírez-Ocampo S., Paniz-Mondolfi A., Rabaan A.A., et al. SARS-CoV, MERS-CoV and now the 2019-novel CoV: have we investigated enough about coronaviruses?–A bibliometric analysis. Trav Med Infect Dis. 2020;33 doi: 10.1016/j.tmaid.2020.101566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rafiei Nasab F., rahim F. 2020.03.19. Bibliometric analysis of global scientific research on SARSCoV-2 (COVID-19) medRxiv. 2020. [Google Scholar]
- 12.Colavizza G., Costas R., Traag V.A., van Eck N.J., van Leeuwen T., Waltman L. 2020.04.20. A scientometric overview of CORD-19. bioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y., Zhou C.C. 2020.05.06. Applying lexical link analysis to discover insights from public information on COVID-19. bioRxiv. 2020. [Google Scholar]
- 14.Wang X., Song X., Guan Y., Li B., Han J. 2020. Comprehensive named entity recognition on cord-19 with distant or weak supervision. arXiv preprint arXiv:200312218. [Google Scholar]
- 15.Dong M., Cao X., Liang M., Li L., Liang H., Liu G. 2020. Understand research hotspots surrounding COVID-19 and other coronavirus infections using topic modeling. medRxiv. [Google Scholar]
- 16.Le Bras P., Gharavi A., Robb D.A., Vidal A.F., Padilla S., Chantler M.J. 2020. Visualising COVID-19 research. arXiv preprint arXiv:200506380. [Google Scholar]
- 17.Älgå A., Eriksson O., Nordberg M. Analysis of scientific publications during the early phase of the COVID-19 pandemic: topic modeling study. J Med Internet Res. 2020;22 doi: 10.2196/21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y.-M., Delen D. Medical informatics research trend analysis: a text mining approach. Health Inf J. 2016 doi: 10.1177/1460458216678443. [DOI] [PubMed] [Google Scholar]
- 19.Miner G., Elder J., Fast A., Hill T., Delen D. Academic Press; 2012. Practical text mining and statistical analysis for non-structured text data applications. [Google Scholar]
- 20.Zengul F.D., Lee T., Delen D., Almehmi A., Ivankova N.V., Mehta T., et al. Research themes and trends in ten top-ranked nephrology journals: a text mining analysis. Am J Nephrol. 2020;51:147–159. doi: 10.1159/000504871. [DOI] [PubMed] [Google Scholar]
- 21.NIH-COVID-19-Portfolio . 2020. NIH COVID-19 portfolio. NIH: NIH. [Google Scholar]
- 22.JMP14. JMP 14 online documentation - basic analysis- text eplorer. SAS Instutute Inc.; 2018. [Google Scholar]
- 23.Evangelopoulos N., Zhang X., Prybutok V.R. Latent semantic analysis: five methodological recommendations. Eur J Inf Syst. 2012;21:70–86. [Google Scholar]
- 24.Landauer T.K., Dumais S.T. A solution to Plato's problem: the latent semantic analysis theory of acquisition, induction, and representation of knowledge. Psychol Rev. 1997;104:211. [Google Scholar]
- 25.Abdi H., Williams L.J., Valentin D. Multiple factor analysis: principal component analysis for multitable and multiblock data sets. Wiley Interdiscipl Rev: Comput Stat. 2013;5:149–179. [Google Scholar]
- 26.Abdi H., Valentin D. Multiple correspondence analysis. Encyclopedia Measure Stat. 2007;2:651–666. [Google Scholar]
- 27.Wiley Lindsay F. Public health law and science in the community mitigation strategy for Covid-19. J Law Biosci. January-June 2020;7(1) doi: 10.1093/jlb/lsaa019. lsaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemel D.J., Rodriguez D.B. Northwestern Public Law Research Paper; 2020:20-05. A public health framework for COVID-19 business liability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Grange L. Covid-19 pandemic and the prospects of education in South Africa. Prospects. 2020:1–12. doi: 10.1007/s11125-020-09514-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staunton Ciara, Swanepoel Carmen, Labuschagine Melodie. Between a rock and a hard place: COVID-19 and South Africa’s response. J Law Biosci. January-June 2020;7(1) doi: 10.1093/jlb/lsaa052. lsaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuller C.D., van Dijk L.V., Thompson R.F., Scott J.G., Ludmir E.B., Thomas C.R. Meeting the challenge of scientific dissemination in the era of COVID-19: toward a modular approach to knowledge-sharing for radiation oncology. Int J Radiat Oncol Biol Phys. 2020;108:496–505. doi: 10.1016/j.ijrobp.2020.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao K.-S., Suryaprakash V., Senthilkumar R., Preethy S., Katoh S., Ikewaki N., et al. Role of immune dysregulation in increased mortality among a specific subset of COVID-19 patients and immune-enhancement strategies for combatting through nutritional supplements. Front Immunol. 2020;11:1548. doi: 10.3389/fimmu.2020.01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaniusas E., Szeles J.C., Kampusch S., Alfageme-Lopez N., Yucumá Conde D., Li X., et al. Non-invasive auricular vagus nerve stimulation as a potential treatment for Covid19-originated acute respiratory distress syndrome. Front Physiol. 2020;11:890. doi: 10.3389/fphys.2020.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayor-Ibarguren A., Robles-Marhuenda Á A hypothesis for the possible role of zinc in the immunological pathways related to COVID-19 infection. Front Immunol. 2020;11:1736. doi: 10.3389/fimmu.2020.01736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q., Allot A., Lu Z. Keep up with the latest coronavirus research. Nature. 2020;579:193. doi: 10.1038/d41586-020-00694-1. [DOI] [PubMed] [Google Scholar]
- 36.Chen Qingyu, Allot Alexis, Lu Zhiyong. LitCovid: an open database of COVID-19 literature. Nucleic Acids Res. 8 January 2021;49(D1):D1534–D1540. doi: 10.1093/nar/gkaa952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.KAGGLE . NIH & The White House. KAGGLE; 2021. COVID-19 open research dataset challenge (CORD-19) an AI challenge with AI2, CZI, MSR, georgetown. [Google Scholar]
- 38.Huong N.Q., Nga N.T.T., Long N.V., Luu B.D., Latinne A., Pruvot M., et al. Coronavirus testing indicates transmission risk increases along wildlife supply chains for human consumption in Viet Nam, 2013-2014. PloS One. 2020;15 doi: 10.1371/journal.pone.0237129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton-Cheh C., Zlotoff D.A., Hung J., Rupasov A., Crowley J.C., Funamoto M. Case 24-2020: a 44-year-old woman with chest pain, dyspnea, and shock. N Engl J Med. 2020;383:475–484. doi: 10.1056/NEJMcpc2004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guagliumi G., Sonzogni A., Pescetelli I., Pellegrini D., Finn A.V. Microthrombi and ST-segment–elevation myocardial infarction in COVID-19. Circulation. 2020;142 doi: 10.1161/CIRCULATIONAHA.120.049294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah P.K., Perez-Iratxeta C., Bork P., Andrade M.A. Information extraction from full text scientific articles: where are the keywords? BMC Bioinf. 2003;4:20. doi: 10.1186/1471-2105-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blei D.M., Ng A.Y., Jordan M.I. Latent dirichlet allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- 43.Anaya L.H. ERIC; 2011. Comparing latent dirichlet allocation and latent semantic analysis as classifiers. [Google Scholar]
- 44.Angelov D. 2020. Top2Vec: distributed representations of topics. arXiv preprint arXiv:200809470. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.