FIGURE 2.
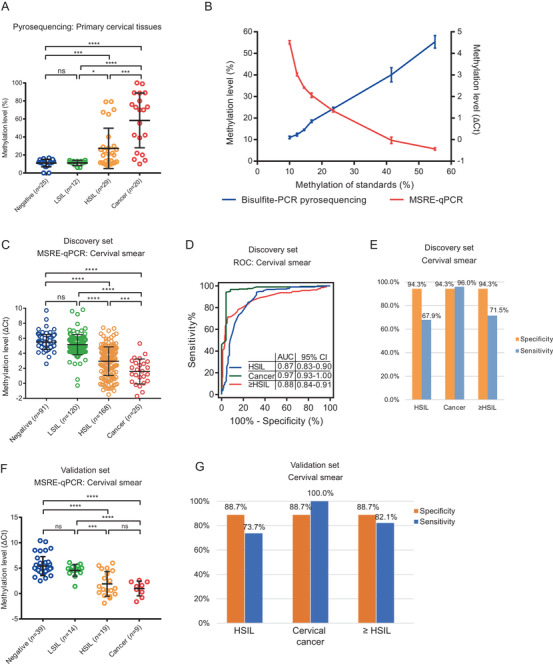
PCDHGB7 was specifically hypermethylated in cervical cancer and HSIL samples. (A) PCDHGB7 methylation level was detected by bisulfite‐PCR pyrosequencing in 86 primary cervical tissue samples. (B) The performance of bisulfite‐PCR (BS‐PCR) pyrosequencing and MSRE‐qPCR in detecting selected DNA methylation standard samples. The x‐axis indicates the DNA methylation level; seven standard samples were detected; the y‐axis in the left indicates the methylation level detected by bisulfite‐PCR pyrosequencing, the y‐axis in the right indicates the ΔCt detected by MSRE‐qPCR, and the ΔCt value reflects the DNA methylation. The repeats of pyrosequencing and MSRE‐qPCR were two and three for each grad, respectively. The mean ± SD values were plotted. (C) PCDHGB7 methylation level of 404 cervical smears in discovery set by MSRE‐qPCR. (D) The ROC curve in 404 cervical smears, and AUC values were illustrated. (E) The sensitivity and specificity of PCDHGB7 hypermethylation in HSIL, CC, and ≥HSIL group in cervical smears in discovery set. (F) PCDHGB7 methylation level of 81 cervical smears in validation set by MSRE‐qPCR. (G) The sensitivity and specificity of PCDHGB7 hypermethylation in HSIL, CC, and ≥HSIL group in cervical smears in validation set. In (A), (C), and (F), error bar represents upper quartile, lower quartile, and median. P values were calculated by the unpaired parametric test with GraphPad Prism 7.0. ns, not significant; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001
