Abstract
Many interactions between microbes and their hosts are driven or influenced by glycans, whose heterogeneous and difficult to characterize structures have led to an underappreciation of their role in these interactions compared to protein-based interactions. Glycans decorate microbe glycoproteins to enhance attachment and fusion to host cells, provide stability, and evade the host immune system. Yet, the host immune system may also target these glycans as glycoepitopes. In this review, we provide a structural perspective on the role of glycans in host-microbe interactions, focusing primarily on viral glycoproteins and their interactions with host adaptive immunity. In particular, we discuss a class of topological glycoepitopes and their interactions with topological mAbs, using the anti-HIV mAb 2G12 as the archetypical example. We further offer our view that structure-based glycan targeting strategies are ready for application to viruses beyond HIV, and present our perspective on future development in this area.
Keywords: glycoepitope, glycans, 2G12, topology, virus
Introduction
Glycosylation is a common post-translational modification of all proteins where complex glycans are attached to specific amino acids such as Asn (in the case of N-linked glycans) and Ser or Thr (in the case of O-linked glycans). Consequently, complex glycans are displayed on surfaces of viruses and host tissues and cells and play multidimensional roles in pathogen-host interactions (Van Breedam et al., 2014; Stroh and Stehle, 2014; Smith and Cummings, 2014; Raman et al., 2014). The structural diversity of these glycans is governed primarily by the complex non-template driven biosynthetic machinery of the host cell involving several enzymes that show tissue-specific expression patterns (Lowe and Marth, 2003; Taylor and Drickamer, 2003; Varki et al., 2008). Additionally, glycan diversity is also influenced by the spatial distribution of the glycosylation sites on the protein surface, wherein glycan clustering can result in steric hindrance of glycan processing enzymes (Bonomelli et al., 2011).
Glycans on host cell or tissue surfaces serve as attachment factors, co-receptors, or primary receptors that are specifically recognized by the viral surface glycoproteins. For example, complex glycans terminated by α2-3- or α2-6-linked sialic acid (N-acetyl neuraminic acid) act as receptors for several different viruses (Stencel-Baerenwald et al., 2014). Linear sulfated glycosaminoglycans such as heparan sulfate act as co-receptors for a variety of viruses (Wadstrom and Ljungh, 1999) including dengue virus (Chen et al., 1997; Artpradit et al. 2013), hepatitis C virus (Baumert et al., 2014), and foot-and-mouth disease virus (Fry et al., 1999). The predominant display of specific glycan motifs on surfaces of different cells and tissues contributes to the host restriction and cell/tissue tropism of the virus (de Vries et al., 2010). As an example, the human upper respiratory epithelial surface predominantly displays sialylated glycan receptors terminated by α2-6-linked sialic acid and these receptors are specifically recognized by hemagglutinin glycoprotein (HA) on the surface of influenza A viruses that are known to infect and transmit via respiratory droplets in humans (Neumann and Kawaoka, 2006; Malik, 2009; Ge and Wang, 2011; Raman et al., 2014). On the other hand, when influenza A viruses that are commensal or epizootic in birds infect humans, they typically affect deep lung and other tissues that predominantly display sialylated glycan receptors terminated by α2-3-linked sialic acid and are unable to transmit efficiently via respiratory droplets. In other cases, host cells also display proteins that specifically bind to glycans on pathogen surfaces thereby acting as key receptors for glycan-mediated viral infection (Figure 1A).
FIGURE 1.
Glycan interactions between host and virus. (A), glycan interactions between host cells and viruses are predominantly mediated by host or virus glycoproteins and their complementary host or virus glycan binding proteins. These binding events trigger processes that favor either viral attachment and fusion or host recognition and response. (B), Hemagglutinin H3 from Influenza A Wyoming/3e/2003, depicted with Man9 glycans and theoretical mAbs for illustrative purposes. Glycans shield vulnerable protein epitopes from neutralizing mAbs, such as the receptor binding site, while non-vulnerable protein epitopes lack selective pressure for glycan shielding. However, these glycans can also present targets as glycoepitopes. Generated in PyMOL from PDB: 6BKN (Wu et al., 2018).
The glycans displayed on viral surfaces are post-translational modifications of viral surface proteins such as envelop proteins in flaviviruses (i.e., dengue and zika viruses) and glycoprotein spikes (i.e., influenza A, Ebola, and SARS-CoV-2 viruses). These glycans are added to the viral glycoproteins as a part of the host-cell glycan biosynthesis during viral replication in the host. In many cases, glycosylation sites on viral surface proteins are highly conserved since the glycans at these sites critically maintain the stability of these proteins and the viral particles as a whole (Wagner et al., 2002; Zhang et al., 2015). In addition to maintaining the stability of the viral particles, these glycans also play key roles in mediating infection of host cells by certain viruses such as dengue and Ebola viruses through specific interactions with glycan-binding proteins (such as C-type lectins) displayed on the host cell surface (Klimstra et al., 2003; Mondotte et al., 2007; Pipirou et al., 2011; Van Breedam et al., 2014).
The complex glycans on the viral surface also play a key role in host immune response to counter the viral infection. While glycan-binding proteins anchored on surfaces of host cells such as dendritic cells (DCs) play a role in viral entry, they also play a dual role to enhance antigen presentation and processing for adaptive immune response (Van Breedam et al., 2014). Sites of N-linked glycosylation are often positively selected during evolution of the virus in human host to increase glycans on the viral surface thereby presenting glycans that mimic self-antigens and mask the underlying protein epitope which in turn permits the virus to evade host immune response. In other cases, particularly with HIV, the clustering of glycosylation sites on the gp120 surface glycoprotein presents novel glycoepitopes that do not mimic self-antigens and therefore lead to potent neutralization by antibodies that target these novel epitopes across a broad spectrum of viral strains (Doores, 2015).
In this perspective, we review the role of viral glycan-host interactions, emphasizing the mechanisms through which glycan structure drives virus immune shielding or immunogenicity. We discuss the role of viral surface glycans in the context of presenting glycoepitopes for the host immune system and examine the various categories of glycoepitopes according to their structure and interaction with glycan-binding agents and antibodies. In particular, we examine a class of high-mannose glycoepitopes we refer to as topological glycoepitopes, using the anti-HIV mAb 2G12 as the archetypical example. Finally, we provide our perspective on fruitful directions for future investigation of topological glycoepitopes, and the rational engineering of antibodies targeting them.
Viral Surface Glycosylation and Host Immune Response
During interactions with host immunity, glycans fall along a spectrum from immunogenic to immune shielding depending on their species, location on the viral glycoprotein, and topological arrangement relative to one another. A major function of N-linked glycosylation on virus glycoproteins is shielding antigenic sites from viruses, as is particularly well established in the cases of Influenza A Tate et al. (2014) and HIV Mascola and Montefiori (2003). This adaptation has a clear structure function-relationship, in which glycans are commonly added adjacent to major functional sites that tend to be the targets of neutralization by host antibodies, thus focusing immune responses away from these sensitive epitopes Bajic et al. (2019), such as for the Flu head proximal to the functional RBS (Figure 1B). In another example, the functional attachment and fusion domains of the Ebola glycoprotein are protected by a glycan cap, which is hypothesized to prevent neutralizing host antibody responses directed against this major functional target (Lee et al., 2008). Similarly, the Epstein-Barr virus is protected by a glycan shield, which has been shown to additionally impair T cell responses directed against infected cells (Gram et al., 2016). The structural basis for these cases of shielding is rather straightforward—glycans sterically prevent host immune recognition of protein epitopes at these sites. Indeed, in a recent investigation of the SARS-CoV-2 glycan shield, Watanabe et al. leveraged extensive site-specific glycan analysis of the glycoproteins of HKU1, SARS, MERS, LASV, H3N2, HIV-1 Env, SIV Env to highlight a correlation between glycoprotein glycan shield density, oligomannose content, and viral immune evasion (Watanabe et al., 2020). Further evidence for this effect can be observed in the viral evolution of Influenza A, which has been shown to add N-glycan sites every 5–7 years Altman et al. (2019) as a result of selective pressure on sensitive protein epitopes in a pattern known as a “glycan clock.” In contrast, glycans decorating less immunogenic regions tend to be conserved, and may contribute beneficially through other mechanisms such as stabilizing the fusion event (Ohuchi et al., 1997).
While immune-shielding glycans are beneficial for immune evasion, they do not solely accumulate over time. Glycosylation bears fitness costs due to increased metabolic resources required for replication and the potential disruption of glycoprotein function. Therefore, immune-shielding glycans may be maintained or eventually lost, often with significant impact on host-virus interaction. Spontaneous glycan loss proximal to the receptor binding site drove virulence of 2009 H1N1 (Wei et al., 2010). This finding was supported by a subsequent study in mice that found evidence that a similar event drove the virulence of the 1918 pandemic H1N1 (Sun et al., 2013). However, these fitness tradeoffs vary significantly across viruses. As compared to the global evolution of Flu, HIV’s glycan shield provides a persistent shielding benefit, both within chronically-infected hosts Coss et al. (2016) and evidently throughout global evolution.
Further, glycan structure and function are dependent on neighboring glycan context for a given glycosylation site. As N-glycan sites accrue in close proximity to one another over the course of viral evolution, their clustering tends to result in oligomannosylation of the N-glycans at nearby sites. The structural mechanisms by which clustered N-glycan sites result in high mannose species have been discussed elsewhere (Behrens and Crispin, 2017). In brief, the effect is likely driven by high glycan density sterically limiting α-mannosidase mediated glycan trimming during glycan processing in the Golgi. This effect appears to be conserved across viruses, and is best defined for HIV in which it is conserved across a range of HIV clades and production systems (Bonomelli et al., 2011). Further, in a recent examination of glycan species heterogeneity for various recombinant Flu vaccine production systems, the FDA found significant glycan variance for the less clustered and non-oligomannose glycans of H1N1 across egg, insect, or eukaryotic cell production systems, yet found that high oligomannose content was preserved across these production systems for the more clustered glycans of H3N2 (An et al., 2019). Still, other mechanisms may also contribute to the presence of high-mannose glycans on certain viruses in certain host contexts. A recent ferret study found that Influenza infection of lung cells induces high-mannose N-glycans on host proteins in a manner dependent on activation of the unfolded protein response (UPR) via IRE1 (Heindel et al., 2020). It is plausible that this pathway may also contribute to oligomannosylation of glycans on viral glycoproteins, and this would likely be independent of N-glycan clusters but rather a property of viral infection itself activating the UPR. Importantly, high-mannose glycans are relatively rare on mature host cells, and are typically only presented within the body at high frequency on stem cell membranes (An et al., 2012). As such, high-mannose glycans are better recognized as foreign by the host, and they themselves may become the target of immune responses, serving as “glycoepitopes.” The canonical example of this recognition occurs on HIV gp120 (Doores et al., 2010), and will be discussed in detail in the following section.
Defining Glycoepitopes on Viral Surfaces
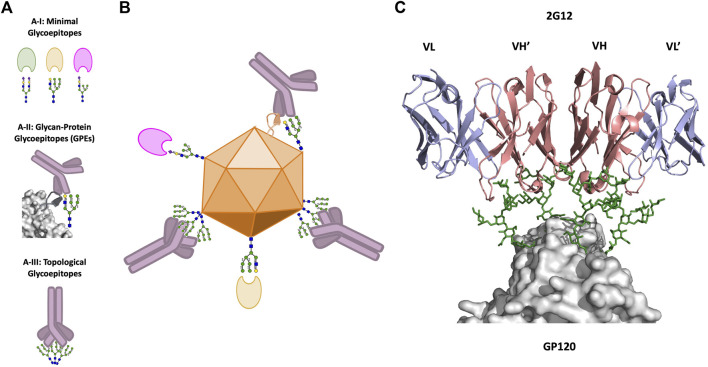
While we have thus far discussed glycans predominantly as beneficial to viral fitness in interactions with the host immune system, glycans may also serve as immune targets (glycoepitopes). We next review three broad types of glycoepitopes, categorized according to the structural-basis for their recognition event (Figure 2A). These types are: 1) minimal glycoepitopes: glycoepitopes recognized chiefly based on their glycan species, typically most effectively by lectins or pattern recognition receptors (Figure 2A-I); 2) glycan-protein glycoepitopes (GPEs): glycoepitopes formed by combinations of glycan and peptide epitopes and recognized by antibodies (Figures 2A-II); and 3) topological glycoepitopes: glycoepitopes occurring at N-glycan clusters with a predisposition for oligomannose, and recognized by unconventional antibodies based on their glycan topology without regard for protein sequence (Figures 2A-III). We focus primarily on the third class, using the anti-HIV mAb 2G12 as the canonical topological mAb to explore the unique features of topological glycoepitopes.
FIGURE 2.
Viral glycoepitopes. (A), Viruses present three types of glycoepitopes for targeting by the host immune system. 2A–I: Minimal glycoepitopes consist of glycans which are effectively recognized specifically by host lectins. 2A–II: Glycoprotein glycoepitopes (GPEs) include both glycan and peptide constituents, and are most effectively neutralized by Y-shaped mAbs. 2A–III: Topological glycoepitopes exist at clusters of high-mannose glycans, and are flexibly recognized by X-shaped mAbs such as 2G12. (B), illustration of the three types of glycoepitopes and their preferred mode of host-recognition. (C), crystal structure of the interaction between the canonical topological glycoepitope on HIV and the X-shaped mAb 2G12 (PDB: 6OZC, Seabright et al., 2020). Heavy chains are shown in red, light chains are shown in blue, glycans are shown in green, and the virus protein surface is shown in gray (PyMol, 2019).
Minimal Glycoepitopes
First, glycoepitopes defined primarily according to the glycan species represents the broadest group, with extent of structural characterization depending on the specific biological context and availability of lectins binding these species, such as mannose-binding lectin and its mannose target (Ji et al., 2005) (Figures 2A-I). These glycoepitopes are recognized by host pattern recognition receptors (PRRs) to activate a variety of host immune responses, typically via opsonization (Brown et al., 2018). These recognition events occur across microbes, including viruses, bacteria, and fungi, and their structural basis has been reviewed extensively elsewhere (Wesener et al., 2017). We note that many agents binding this class of glycoepitope have significant recognition breadth, and may also engage the following two groups of glycoepitopes, albeit usually with reduced specificity and affinity, which has tended to complicate therapeutic development of these agents. Recent engineering efforts have sought to optimize these properties in pursuit of enhanced clinical applicability Hamorsky et al. (2019); Covés-Datson et al. (2020), and thus represent a creative and orthogonal approach to the GPE and topological glycoepitope targeting strategies we discuss next.
Glycan-Protein Glycoepitopes
Second, glycoepitopes may be defined by a combination of glycan and protein, with targeting primarily driven by antibodies (Figures 2A-II). Human IgG and IgM serum have high concentrations of anti-glycan antibodies targeting a diverse range of carbohydrate antigens, including antibodies against both xeno-antigens [e.g. Gal(\alpha1-3)Gal] Bovin et al. (2012) and allo-antigens (e.g., ABO blood group associated antigens) (Yamamoto et al., 1990). Many of these antibodies recognize di- or tri-saccharide motifs, but fail to recognize the same motif in the context of a longer complex glycan, a masking effect that is hypothesized to limit autoimmune response Bovin et al. (2012), although this effect cannot necessarily be attributed to size alone (Huflejt et al., 2009).
Anti-glycan antibodies can be divided into natural and antigen-dependent antibodies. Natural antibodies exist without any antigenic stimulation or T-Cell dependence, while antigen-dependent antibodies are generated and matured specifically in response to antigenic stimuli. The mechanism of antigen-dependent immune response to various glycoepitopes (glycoprotein, glycolipid, oligosaccharide) is not fully understood, due to their heterogeneous nature and their relationship to the well-established T-cell response for peptide antigens (Kappler and Hennet, 2020). However, research into the subset of antibodies targeting glycan-protein epitopes (GPEs) on glycoproteins has provided structural evidence of antigen-mediated antibody evolution. Identifying the critical structural mechanisms of binding for these GPE targeting mAbs can be used to optimize both therapeutic mAb design and vaccine design.
Immune response to the HIV env glycoprotein, due to its extensive glycan shield, has generated broadly neutralizing HIV antibodies targeting several distinct glycoepitopes (Burton et al., 2009; Pejchal et al., 2011; Walker et al., 2011; Sanders et al., 2013; Falkowska et al., 2014). The glycan-dependence of these antibodies varies from fully glycan-dependent with no associated protein interface (2G12, discussed in the next sub-section on topological glycoepitopes), to protein and glycan-dependent (PG9, PG16, and PGT121-128) (Burton et al., 2009; Pejchal et al., 2010). Examining the similarities and differences of GPE mAbs (“PGT121-124”) (Walker et al., 2011; Garces et al., 2014) illustrates key aspects of canonical paratope-GPE interaction: broadly similar paratope structure, divergent mechanisms of interaction after affinity maturation, and glycan-species dependent binding.
Antibodies PGT121-124 all share a canonical paratope structure that has properties characteristic of GPE-binding mAbs. An abnormally-long H3 loop in association with the light chain forms an elongated face that interacts with the GPE glycan at residue N332 (Julien et al., 2013), and a “GDIR” motif (residues 324-327) that interacts with the protein components of the GPE (Garces et al., 2014). Further, the interactions with GDIR and N332 are well-defined in all the mAbs in this lineage and fixed early in the maturation process from germline (Garces et al., 2015). That is, a long H3 loop to penetrate the glycan shield while also interacting with the GPE glycan, a peptide-binding motif, and a stabilizing glycan are critical attributes of this epitope-paratope interaction that may characterize other GPE epitope-paratope interactions as well. PGT121-124 have similar high-level structural characteristics, but differ significantly in terms of their final paratope amino acid sequences due to diverging paths of natural affinity maturation. In addition to the “elongated face,” the PGT121-124 also contain an “open face” that is comprised of residues in CDRH1, CDRH2, CDRH3 (Garces et al., 2014). Somatic hypermutation during affinity maturation in this open face leads to the final differences between antibodies. Interestingly, this affinity maturation optimizes interaction with an additional glycan at N137 Garces et al. (2015), showing that affinity maturation against glycan species is indeed possible from a germline with the characteristics to bind a GPE.
Glycan recognition by GPE targeting antibodies can be highly specific, even within closely related antibodies. Despite originating from the same putative germline, related PGT121-123 and “10-1074-like” antibodies exhibit distinct binding sensitivity to glycan modulation. PGT121-123 antibodies were more affected after PNGase F treatment, while 10-1074-like antibodies were more affected by Endo H treatment, indicating a preference of PGT121-123 for complex glycans and 10-1074 like antibodies for high-mannose species (Mouquet et al., 2012). Additionally, PGT121-123 only bound to complex, and not high-mannose, glycans on a microarray, while 10-1074-like antibodies did not bind to any glycans without protein (Mouquet et al., 2012). This evidence, in combination with evidence that GPE-Abs undergo glycan-directed affinity maturation, suggests that GPE targeting antibodies are sensitive to the glycan species. However, evaluation of the species-specificity is challenging due to the interdependence of the paratope-peptide binding, the paratope-glycan binding, and potential stabilizing or structure altering effects of the GPE in the context of the full system.
Topological Glycoepitopes
The third group of glycoepitopes are topological glycoepitopes, which are fully glycan-dependent without a protein component (Figures 2A-III). Instead, topological glycoepitopes exist at clusters of high-mannose glycans that are spatially distributed in a distinct topological arrangement on the viral surface. The high-mannose type and the spatial arrangement of these glycans on the viral surface present distinct topological glycoepitopes that are recognized by neutralizing antibodies. The most extensively characterized example of topological glycoepitopes is that of the anti-HIV antibody 2G12 on the surface of the gp120 protein. However, topological glycoepitopes do not appear unique to HIV, and recent evidence suggests that 2G12 recognizes topological glycoepitopes on additional viruses. At the time of this writing, 2G12 glycoepitopes have been identified on Influenza H3N2 Lee et al. (2021) and SARS-CoV-2 (Acharya et al., 2020; Mannar et al., 2021).
The recognition of a topological glycoepitope which comprised exclusively of spatially distributed glycans was first reported upon discovery of the 2G12 antibody in 1996 (Trkola et al., 1996). 2G12 is a domain-exchanged (X-shaped) mAb whose format presents three glycan binding surfaces (VH-VL’, VH’-VL, and VH-VH’) (Figure 2, Right). While glycan-Ab binding events tend to be relatively low affinity, the three binding surfaces uniquely allow 2G12 to engage clusters of high-mannose N-glycans in a multivalent and high-affinity interaction (Scanlan et al., 2002; Calarese et al., 2003). The 2G12 binding interface has been investigated intensively, with multiple structures for 2G12 bound to individual glycans Sanders et al. (2013), lipooligosaccharides Stanfield et al. (2015), and also to recombinant HIV Env trimer BG505 SOSIP.664 (Seabright et al., 2020). These studies, via binding competition experiments, indicate a binding preference for terminal α1-2 linked-mannose on the Man9 d1 arm, but demonstrate binding albeit with reduced affinity to d2 and d3 arms, as well as to terminal mannose on Man1-Man8 (Sanders et al., 2002; Calarese et al., 2005; Dunlop et al., 2010; Seabright et al., 2020). These data indicate that 2G12’s glycan binding surfaces are highly accommodative of variable mannose content, which is likely a key feature of topological mAbs, given that mannose content may vary across a given topological glycoepitope.
Topological mAbs exhibit flexible valency within topological glycoepitopes. The two most recent and highest quality structural investigation of the 2G12-gp120 interaction indicate that 2G12 binds high-mannose glycans at sites N295, N332, N339, and N392 (Murin et al., 2014; Seabright et al., 2020). The investigation by Seabright et al. included a site-specific glycan knockout experiment for glycans within and adjacent to the 2G12 glycoepitope, finding a high degree of 2G12 binding flexibility. Knockout of any one of the four binding glycans only reduced 2G12 binding between two- and five-fold, with the greatest reduction still maintaining nanomolar binding affinity (17 nM; Seabright et al., 2020). Meanwhile, the Murin study conducted a similar glycan knockout experiment, using pseudovirus neutralization rather than binding as the readout (Murin et al., 2014). Their results indicate that glycans at sites 295, 332, and 392 are critical for pseudoviral neutralization. As is being increasingly appreciated for mAbs targeting protein epitopes, subtle epitope changes can drive critical variations in mAb function despite minor changes in mAb binding strength. This pair of binding and neutralization knock-out experiments from Seabright et al. and Murin et al. indicate that the same may be true for topological mAbs and their glycoepitopes.
The glycans within topological glycoepitopes display significant higher-order network effects. The knock-out studies performed by Seabright et al. demonstrate that N-glycans residing outside of the 2G12 binding residues affect 2G12 binding (Seabright et al., 2020). This effect is realized primarily through two mechanisms: 1) adjacent glycans contribute to the degree of N-glycan clustering and thus influence the degree of oligomannosylation at the binding glycans, and 2) adjacent glycans interact with and stabilize or destabilize the binding glycans. These properties of glycan clusters emphasize why topological glycoepitopes must be defined in the context of their glycan network, and certainly beg the question “where do topological glycoepitopes begin and end?.”
Topological glycoepitopes can form on quaternary structures, and this quality may contribute to the neutralizing ability of antibodies targeting topological glycoepitopes. Glycan clustering is amplified at the quaternary junctions formed by protein oligomers, such as trimeric viral proteins like SARS-CoV-2 spike protein and Influenza HA. Indeed, Lee et al. provide evidence that the 2G12 glycoepitope on Influenza H3N2 is quaternary and neutralizing, though the neutralizing effect may also be driven by the glycoepitope’s location proximal to the functionally-important receptor binding site (Lee et al., 2021). The 2G12 topological glycoepitope on SARS-CoV-2 is also quaternary (Acharya et al., 2020). Targeting quaternary epitopes has become increasingly appreciated as an effective neutralization strategy. Quaternary protein epitopes have been characterized for a number of viruses including HIV Robinson et al. (2010) and Zika Tharakaraman et al. (2018); Long et al. (2019), and shown to contribute to high affinity binding and potent neutralization due to inter-chain locking. Quaternary epitopes have also been shown to mediate differential and beneficial effects for mAbs targeting CD20, in which binding an epitope spanning a single tetramer vs. an epitope cross-linking adjacent tetramers results in a differential mechanism of cell killing (Marshall et al., 2017; Meyer et al., 2018). Recently, a linked biparatopic nanobody-pair that crosslinks adjacent SARS-CoV-2 Spike protein monomers was shown to have a synergistic neutralization effect as compared to the two nanobodies in combination but not linked (Koenig et al., 2021). Thus, targeting quaternary epitopes is of particular interest for imparting enhanced functionality during antibody design, and topological mAbs may be uniquely suited to target quaternary epitopes because they have no direct dependency on the protein surface at these junctions.
Structural Characterization and Modeling of Topological Glycoepitopes
Topological glycoepitopes present unique challenges for targeting and antibody engineering. Topological glycoepitopes exist on the surfaces of a network of mobile high-mannose glycans, and so projection angles, heterogeneity, and flexibility complicate rigid structural definition that traditionally serves as the basis for most epitope-paratope complexes. Despite a previous statistical analysis of glycoproteins in the PDB providing evidence that glycan structures on homologous proteins are homogenous, the same study found that glycan orientation with respect to the protein surface was highly variable (Jo et al., 2013). Alternatively, more recent studies of the conformational heterogeneity of smaller carbohydrates (trisaccharides) demonstrate that computing the full range of conformations is required to reconcile predictions with experimental results at the atomic level (Yang et al., 2016). Dr. Robert Woods famously advised that the question “What is the shape of my glycan?” be rephrased to “What shapes can the glycan adopt?” (Woods and Tessier, 2010). For topological glycoepitopes, this line of questioning must be expanded to include additional parameters that reflect orientation and range of motion relative to the protein surface as well as interactions between glycans. Topological glycoepitope prediction, therefore, should consider the range of sampled conformations rather than a rigid snapshot of a topological mAb-topological glycoepitope complex.
This problem is most relevant for applying computationally-based rational antibody engineering to antibodies targeting topological glycoepitopes, thus requiring an in-silico characterization that models complex glycan-protein systems. Several webservers and computational packages exist specifically for this purpose, including GlyCAM-Web Kirschner et al. (2008), CHARMM-GUI Park et al. (2019), and Glycosylator Lemmin and Soto, (2019). In addition, several complete computational modeling programs such as Rosetta Das and Baker, (2008) include functionality for building and manipulating glycans. Each of these methods relies on a force field approach to predict the optimal (energy minimized) conformation of the system. Although different force fields contain a largely similar set of component energy terms, contributions from various factors (e.g., electrostatic vs van der Waals) are differentially weighted between models, often with the option of user-defined re-weighting which requires significant knowledge of the force field in question (Shirts et al., 2017). Therefore, when run on complex systems that include multiple interacting glycans on a protein surface, algorithms such as GLYCAM-Web and CHARMM-GUI, which rely on the GLYCAM Kirschner et al. (2008) and CHARMM Guvench et al. (2011) force fields respectively, can produce divergent results. In conclusion, unlike in other protein-protein interaction problems where static representations of protein surfaces lead to a significant number of promising docking algorithms Pagadala et al. (2017), the variability of glycoprotein structure predictions makes a single predicted set of atomic coordinates an unsuitable starting point for describing topological glycoepitopes.
Molecular Dynamics (MD) simulations, also based on force fields including those above, have been used to simulate the possible conformations of glycoproteins Lee et al. (2015), including the dynamics of the glycoepitope-containing HIV Env protein (Lemmin et al., 2017; Yang et al., 2017; Ferreira et al., 2018; Berndsen et al., 2020). These studies have elucidated insights regarding the full conformational space of Env glycans, with sampling volumes up to 50000 A3 for Man9 glycans Yang et al. (2017), and are able to reproduce 3D variance observed cryo-EM maps (Berndsen et al., 2020). Despite these promising results, MD simulations are computationally expensive and thus characterizing glycoepitopes across PDB using these methods remains computationally elusive, albeit highly desirable. Shortcuts to accomplish this screening task may be sufficient, such as employing glycosylation site topology as a proxy for glycan topology. Still, more advanced models may be required if rigorous relationships between antibody affinity and glycoepitope properties are to be established, especially toward the goal of rational topological antibody engineering.
Glycoepitopes as Novel Antiviral Targets: A Perspective
Topological mAbs targeting high-mannose topological glycoepitopes have seen little development, yet offer promise as therapeutics, diagnostics, and analytical tools. Two phase one trials provide evidence that 2G12 is a clinically developable template. In the first trial, 2G12 was successfully produced in CHO cells and proven safe when co-administered in combination with another mAb (2F5) to HIV patients (Armbruster et al., 2002). In the second, 2G12 was produced in tobacco plants and shown to be safe during topical administration as a prophylactic for healthy women (Ma et al., 2015). These encouraging early data indicate that application of 2G12 to other viruses, as well as engineering efforts using 2G12 as a template, may prove fruitful.
Rational engineering efforts to expand the scope of topological mAbs may take a variety of forms. Most simply, 2G12 may be modifiable to adjust its topological footprint or glycan binding affinity and specificity, for example through hinge length alterations or paratope engineering. Second, other glycan-binding mAbs may be convertible to domain-exchanged format, which may or may not be sufficient to impart topological qualities. Investigations into the structural mechanism of 2G12 domain exchange identified that just 5-7 substitutions are required to mediate domain exchange Huber et al. (2010), though additional engineering efforts may be required to optimize placement of glycan-binding domains relative to the three topological binding surfaces. We highlight the potential for a screen searching for novel chimeric topological mAbs, via grafting of known glycan-binding CDRs onto the 2G12 scaffold or by induction of domain exchange via mutation of the cross-over residues. In particular, anti-Bacterial glycan-targeting mAbs which tend to have desirable qualities—flexible recognition of multiple species due to targeting of “minimal” glycoepitopes Rollenske et al. (2018)—as well as diagnostic microarrays, which have already characterized sets of mAbs and their glycan specificities Campanero-Rhodes et al. (2020), may serve as logical starting points. As described earlier, computational methods to predict glycan cluster topology and variability would greatly enhance these efforts. Third, such optimizations may also investigate the dimeric form of 2G12, which was shown to increase neutralization potency against HIV by 50-80-fold (West et al., 2009). Subsequent structural investigations suggest that enhanced potency is driven by increased flexibility Wu et al. (2013), and further functional investigation found that the dimeric 2G12 format also enhances ADCC (Klein et al., 2010).
Pursuing topological glycoepitope and topological mAb research efforts could prove particularly useful for eventual rapid response deployment. In one hypothetical scenario, with precedent according to the previously described Flu “glycan clock,” the emergence of an Influenza H1N1 or SARS-CoV-2 variant with a novel N-glycan site obscuring an immunodominant epitope adjacent to the H1N1 RBS or SARS-CoV-2 RBD might result in a sudden increase in virulence in vulnerable populations. In such a scenario, pre-existing development work on 2G12 or other topological mAb rational engineering might prove highly beneficial for rapid response, perhaps via prophylactic administration to vulnerable populations. The need for template engineering, rather than use of WT 2G12, is highlighted by the early data presented earlier Acharya et al. (2020) demonstrating that WT 2G12 glycoepitopes exist on SARS-CoV-2 but do not facilitate neutralization, likely due to their location distal from the spike protein receptor binding domain (RBD). If additional N-glycan sites were to be evolved within the SARS-CoV-2 RBD, a topological mAb might need to be engineered to display a preference for the RBD glycan topology rather than the RBD-distal topological glycoepitope.
In addition to their therapeutic value, topological mAbs could be employed as diagnostic and analytical tools. Due to the difficulty of characterizing glycans and glycoproteins, glycan arrays have historically been deployed to assay samples using sets of glycan binding agents with well-characterized targets. In example, a straightforward application of 2G12 toward this purpose is its use to characterize gp120 or HIV immunogens for glycan presentation (Doores et al., 2013). This has been a mainstay approach to HIV vaccine development, though glycan mimicry alone has thus far fallen short of generating neutralizing responses (Seabright et al., 2019). A similar approach leveraging topological binding to quaternary epitopes could be taken to assay for proper oligomerization of glycoproteins that only display functional topological epitopes in their oligomerized state, for example on Influenza H3N2. Such an analytical tool could prove useful for validating seasonally-adjusted influenza vaccines, integrating well with existing mass spectrometry-based approaches. In conclusion, we believe that topological glycoepitopes offer fruitful therapeutic, diagnostic, and analytical opportunities, and hope that this perspective motivates further study of this interesting epitope-paratope interaction.
Author Contributions
NM, TC, and RR wrote the manuscript; RS supervised and reviewed the work; all authors approved the manuscript for publication.
Funding
NM was supported in part by T32 ES007020/ES/NIEHS NIH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Acharya P., Wilton W., Rory H., Katarzyna J., Kartik M., Robert P., et al. (2020). A Glycan Cluster on the SARS-CoV-2 Spike Ectodomain Is Recognized by Fab-Dimerized Glycan-Reactive Antibodies. bioRxiv 06 (30), 178897. 10.1101/2020.06.30.178897 [DOI] [Google Scholar]
- Altman M. O., Angel M., Košík I., Trovão N. S., Zost S. J., Gibbs J. S., et al. (2019). Human Influenza A Virus Hemagglutinin Glycan Evolution Follows a Temporal Pattern to a Glycan Limit. mBio 10 (2), e00204–19. 10.1128/mBio.00204-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H. J., Gip P., Kim J., Wu S., Park K. W., McVaugh C. T., et al. (2012). Extensive Determination of Glycan Heterogeneity Reveals an Unusual Abundance of High Mannose Glycans in Enriched Plasma Membranes of Human Embryonic Stem Cells. Mol. Cel Proteomics 11 (4), M111.010660. 10.1074/mcp.M111.010660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y., Parsons L. M., Jankowska E., Melnyk D., Joshi M., Cipollo J. F. (2019). N-glycosylation of Seasonal Influenza Vaccine Hemagglutinins: Implication for Potency Testing and Immune Processing. J. Virol. 93 (2), e01693–18. 10.1128/JVI.01693-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster C., Stiegler G. M., Vcelar B. A., Jäger W., Michael N. L., Vetter N., et al. (2002). A Phase I Trial with Two Human Monoclonal Antibodies (hMAb 2F5, 2G12) against HIV-1. AIDS 16 (2), 227–233. 10.1097/00002030-200201250-00012 [DOI] [PubMed] [Google Scholar]
- Artpradit C., Robinson L. N., Gavrilov B. K., Rurak T. T., Ruchirawat M., Sasisekharan R. (2013). Recognition of Heparan Sulfate by Clinical Strains of Dengue Virus Serotype 1 Using Recombinant Subviral Particles. Virus. Res. 176, 69–77. 10.1016/j.virusres.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G., Maron M. J., Adachi Y., Onodera T., McCarthy K. R., McGee C. E., et al. (2019). Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe 25 (6), 827–835.e6. 10.1016/j.chom.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumert T. F., Meredith L., Ni Y., Felmlee D. J., McKeating J. A., Urban S. (2014). Entry of Hepatitis B and C Viruses - Recent Progress and Future Impact. Curr. Opin. Virol. 4, 58–65. 10.1016/j.coviro.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Behrens A. J., Crispin M. (2017). Structural Principles Controlling HIV Envelope Glycosylation. Curr. Opin. Struct. Biol. 44, 125–133. 10.1016/j.sbi.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen Z. T., Chakraborty S., Wang X., Cottrell C. A., Torres J. L., Diedrich J. K., et al. (2020). Visualization of the HIV-1 Env Glycan Shield across Scales. Proc. Natl. Acad. Sci. U S A. 117, 28014. 10.1073/pnas.2000260117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomelli C., Doores K. J., Dunlop D. C., Thaney V., Dwek R. A., Burton D. R., et al. (2011). The Glycan Shield of HIV Is Predominantly Oligomannose Independently of Production System or Viral Clade. PLoS One 6 (8), e23521. 10.1371/journal.pone.0023521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovin N., Obukhova P., Shilova N., Rapoport E., Popova I., Navakouski M., et al. (2012). Repertoire of Human Natural Anti-glycan Immunoglobulins. Do We Have Auto-Antibodies? Biochim. Biophys. Acta 1820 (9), 1373–1382. 10.1016/j.bbagen.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Brown G. D., Willment J. A., Whitehead L. (2018). C-type Lectins in Immunity and Homeostasis. Nat. Rev. Immunol. 18 (6), 374–389. 10.1038/s41577-018-0004-8 [DOI] [PubMed] [Google Scholar]
- Burton D. R., Walker L. M., Phogat S. K., Chan-Hui P. Y., Wagner D., Phung P., et al. (2009). Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science 326, 285. 10.1126/science.1178746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese D. A., Lee H. K., Huang C. Y., Best M. D., Astronomo R. D., Stanfield R. L., et al. (2005). Dissection of the Carbohydrate Specificity of the Broadly Neutralizing Anti-HIV-1 Antibody 2G12. Proc. Natl. Acad. Sci. U S A. 102 (38), 13372–13377. 10.1073/pnas.0505763102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese D. A., Scanlan C. N., Zwick M. B., Deechongkit S., Mimura Y., Kunert R., et al. (2003). Antibody Domain Exchange Is an Immunological Solution to Carbohydrate Cluster Recognition. Science 300 (5628), 2065–2071. 10.1126/science.1083182 [DOI] [PubMed] [Google Scholar]
- Campanero-Rhodes M. A., Palma A. S., Menéndez M., Solís D. (2020). Microarray Strategies for Exploring Bacterial Surface Glycans and Their Interactions with Glycan-Binding Proteins. Front. Microbiol. 10, 2909. 10.3389/fmicb.2019.02909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Maguire T., Hileman R. E., Fromm J. R., Esko J. D., Linhardt R. J., et al. (1997). Dengue Virus Infectivity Depends on Envelope Protein Binding to Target Cell Heparan Sulfate. Nat. Med. 3 (8), 866–871. 10.1038/nm0897-866 [DOI] [PubMed] [Google Scholar]
- Coss K. P., Vasiljevic S., Pritchard L. K., Krumm S. A., Glaze M., Madzorera S., et al. (2016). HIV-1 Glycan Density Drives the Persistence of the Mannose Patch within an Infected Individual. J. Virol. 90 (24), 11132–11144. 10.1128/JVI.01542-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covés-Datson E. M., King S. R., Legendre M., Gupta A., Chan S. M., Gitlin E., et al. (2020). A Molecularly Engineered Antiviral Banana Lectin Inhibits Fusion and Is Efficacious against Influenza Virus Infection In Vivo . Proc. Natl. Acad. Sci. U.S.A 117 (4), 2122–2132. 10.1073/pnas.1915152117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., Baker D. (2008). Macromolecular Modeling with Rosetta. Annu. Rev. Biochem. 77, 363. 10.1146/annurev.biochem.77.062906.171838 [DOI] [PubMed] [Google Scholar]
- de Vries R. P., de Vries E., Bosch B. J., de Groot R. J., Rottier P. J., de Haan C. A. (2010). The Influenza A Virus Hemagglutinin Glycosylation State Affects Receptor-Binding Specificity. Virology 403 (1), 17–25. 10.1016/j.virol.2010.03.047 [DOI] [PubMed] [Google Scholar]
- Doores K. J., Bonomelli C., Harvey D. J., Vasiljevic S., Dwek R. A., Burton D. R., et al. (2010). Envelope Glycans of Immunodeficiency Virions Are Almost Entirely Oligomannose Antigens. Proc. Natl. Acad. Sci. U S A. 107 (31), 13800–13805. 10.1073/pnas.1006498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K. J., Huber M., Le K. M., Wang S. K., Doyle-Cooper C., Cooper A., et al. (2013). 2G12-expressing B Cell Lines May Aid in HIV Carbohydrate Vaccine Design Strategies. J. Virol. 87 (4), 2234–2241. 10.1128/JVI.02820-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K. J. (2015). The HIV Glycan Shield as a Target for Broadly Neutralizing Antibodies. FEBS J. 282, 4679–4691. 10.1111/febs.13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop D. C., Bonomelli C., Mansab F., Vasiljevic S., Doores K. J., Wormald M. R., et al. (2010). Polysaccharide Mimicry of the Epitope of the Broadly Neutralizing Anti-HIV Antibody, 2G12, Induces Enhanced Antibody Responses to Self Oligomannose Glycans. Glycobiology 20 (7), 812–823. 10.1093/glycob/cwq020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E., Le K. M., Ramos A., Doores K. J., Lee J. H., Blattner C., et al. (2014). Broadly Neutralizing HIV Antibodies Define a Glycan-dependent Epitope on the Prefusion Conformation of Gp41 on Cleaved Envelope Trimers. Immunity 40 (5), 657–668. 10.1016/j.immuni.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R. C., Grant O. C., Moyo T., Dorfman J. R., Woods R. J., Travers S. A., et al. (2018). Structural Rearrangements Maintain the Glycan Shield of an HIV-1 Envelope Trimer after the Loss of a Glycan. Scientific Rep. 8, 15031. 10.1038/s41598-018-33390-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry E. E., Lea S. M., Jackson T., Newman J. W., Ellard F. M., Blakemore W. E., et al. (1999). The Structure and Function of a Foot-And-Mouth Disease Virus-Oligosaccharide Receptor Complex. Embo J. 18, 543–554. 10.1093/emboj/18.3.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F., Lee J. H., de Val N., Torrents de la Pena A., Kong L., Puchades C., et al. (2015). Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity 43, 1053. 10.1016/j.immuni.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F., Sok D., Kong L., McBride R., Kim H. J., Saye-Francisco K. F., et al. (2014). Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies. Cell. 159 (1), 69–79. 10.1016/j.cell.2014.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S., Wang Z. (2011). An Overview of Influenza A Virus Receptors. Crit. Rev. Microbiol. 37, 157–165. 10.3109/1040841X.2010.536523 [DOI] [PubMed] [Google Scholar]
- Gram A. M., Oosenbrug T., Büll C., Comvalius A., Dickson K. J. I., et al. (2016). The Epstein-Barr Virus Glycoprotein Gp150 Forms an Immune-Evasive Glycan Shield at the Surface of Infected Cells. Plos Pathog. 12 (4), e1005550. 10.1371/journal.ppat.1005550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvench O., Mallajosyula S. S., Raman E. P., Hatcher E., Vanommeslaeghe K., Foster T. J., et al. (2011). CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and its Utility in Polysaccharide and Carbohydrate-Protein Modeling. J. Chem. Theor. Comput. 7, 3162. 10.1021/ct200328p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamorsky K. T., Kouokam J. C., Dent M. W., Grooms T. N., Husk A. S., Hume S. D., et al. (2019). Engineering of a Lectibody Targeting High-mannose-type Glycans of the HIV Envelope. Mol. Ther. 27 (11), 2038–2052. 10.1016/j.ymthe.2019.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel D. W., Koppolu S., Zhang Y., Kasper B., Meche L., Vaiana C. A., et al. (2020). Glycomic Analysis of Host Response Reveals High Mannose as a Key Mediator of Influenza Severity. Proc. Natl. Acad. Sci. U.S.A 117 (43), 26926–26935. 10.1073/pnas.2008203117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M., Le K. M., Doores K. J., Fulton Z., Stanfield R. L., Wilson I. A., et al. (2010). Very Few Substitutions in a Germ Line Antibody Are Required to Initiate Significant Domain Exchange. J. Virol. 84 (20), 10700–10707. 10.1128/JVI.01111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huflejt M. E., Vuskovic M., Vasiliu D., Xu H., Obukhova P., Shilova N., et al. (2009). Anti-carbohydrate Antibodies of normal Sera: Findings, Surprises and Challenges. Mol. Immunol. 46, 3037. 10.1016/j.molimm.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Ji X., Gewurz H., Spear G. T. (2005). Mannose Binding Lectin (MBL) and HIV. Mol. Immunol. 42 (2), 145–152. 10.1016/j.molimm.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Jo S., Lee H. S., Skolnick J., Im W. (2013). Restricted N-Glycan Conformational Space in the PDB and its Implication in Glycan Structure Modeling. Plos Comput. Biol. 9 (3), e1002946. 10.1371/journal.pcbi.1002946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J. P., Sok D., Khayat R., Lee J. H., Doores K. J., Walker L. M., et al. (2013). Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 Gp120 V3 Base and Multiple Surrounding Glycans. Plos Pathog. 9 (5), e1003342. 10.1371/journal.ppat.1003342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler K., Hennet T. (2020). Emergence and Significance of Carbohydrate-specific Antibodies. Genes Immun. 21 (4), 224–239. 10.1038/s41435-020-0105-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner K. N., Yongye A. B., Tschampel S. M., Daniels C. R., Foley B. L., Woods R. J. J. (2008). Comput. Chem. 29, 622–655. 10.1002/jcc.20820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J. S., Webster A., Gnanapragasam P. N., Galimidi R. P., Bjorkman P. J. (2010). A Dimeric Form of the HIV-1 Antibody 2G12 Elicits Potent Antibody-dependent Cellular Cytotoxicity [published Correction Appears in AIDS. 2010;24(16):2601]. AIDS 24 (11), 1633–1640. 10.1097/qad.0b013e32833ad8c8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra W. B., Nangle E. M., Smith M. S., Yurochko A. D., Ryman K. D. (2003). DC-SIGN and L-SIGN Can Act as Attachment Receptors for Alphaviruses and Distinguish between Mosquito Cell- and Mammalian Cell-Derived Viruses. J. Virol. 77 (22), 12022–12032. 10.1128/jvi.77.22.12022-12032.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P. A., Das H., Liu H., Kümmerer B. M., Gohr F. N., Jenster L. M., et al. (2021). Structure-guided Multivalent Nanobodies Block SARS-CoV-2 Infection and Suppress Mutational Escape. Science 371, eabe6230. 10.1126/science.abe6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. D., Watanabe Y., Wu N. C., Han J., Kumar S., Pholcharee T., et al. (2021). A Cross-Neutralizing Antibody between HIV-1 and Influenza Virus. Plos Pathog. 17 (3), e1009407. 10.1371/journal.ppat.1009407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. S., Jo S., Mukherjee S., Park S. J., Skolnick J., Lee J., et al. (2015). GS-align for Glycan Structure Alignment and Similarity Measurement. Bioinformatics 31 (16), 2653–2659. 10.1093/bioinformatics/btv202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Fusco M. L., Hessell A. J., Oswald W. B., Burton D. R., Saphire E. O. (2008). Structure of the Ebola Virus Glycoprotein Bound to an Antibody from a Human Survivor. Nature 454 (7201), 177–182. 10.1038/nature07082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmin T., Soto C. (2019). Glycosylator: A Python Framework for the Rapid Modeling of Glycans. BMC Bioinformatics 20, 513. 10.1186/s12859-019-3097-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmin T., Soto C., Stuckey J., Kwong P. D. (2017). Microsecond Dynamics and Network Analysis of the HIV-1 SOSIP Env Trimer Reveal Collective Behavior and Conserved Microdomains of the Glycan Shield. Structure 25, 1631. 10.1016/j.str.2017.07.018 [DOI] [PubMed] [Google Scholar]
- Long F., Doyle M., Fernandez E., Miller A. S., Klose T., Sevvana M., et al. (2019). Structural Basis of a Potent Human Monoclonal Antibody against Zika Virus Targeting a Quaternary Epitope. Proc. Natl. Acad. Sci. U S A. 116 (5), 1591–1596. 10.1073/pnas.1815432116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J. B., Marth J. D. (2003). A Genetic Approach to Mammalian Glycan Function. Annu. Rev. Biochem. 72, 643–691. 10.1146/annurev.biochem.72.121801.161809 [DOI] [PubMed] [Google Scholar]
- Ma J. K., Drossard J., Lewis D., Altmann F., Boyle J., Christou P., et al. (2015). Regulatory Approval and a First-In-Human Phase I Clinical Trial of a Monoclonal Antibody Produced in Transgenic Tobacco Plants. Plant Biotechnol. J. 13 (8), 1106–1120. 10.1111/pbi.12416 [DOI] [PubMed] [Google Scholar]
- Mannar D., Leopold K., Subramaniam S. (2021). Glycan reactive anti‐HIV‐I antibodies bind the SARS‐CoV‐2 spike protein but not block viral entry. Available at: www.biorxiv.org/content/10.1101/2021.01.03.425141v1.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik P. J. S. (2009). Avian Influenza Viruses in Humans. Revue scientifique Tech. 28, 161–173. 10.20506/rst.28.1.1871 [DOI] [PubMed] [Google Scholar]
- Marshall M. J. E., Stopforth R. J., Cragg M. S. (2017). Therapeutic Antibodies: What Have We Learnt from Targeting CD20 and where Are We Going?. Front. Immunol. 8, 1245. 10.3389/fimmu.2017.01245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J. R., Montefiori D. C. (2003). HIV-1: Nature's Master of Disguise. Nat. Med. 9 (4), 393–394. 10.1038/nm0403-393 [DOI] [PubMed] [Google Scholar]
- Meyer S., Evers M., Jansen J. H. M., Buijs J., Broek B., Reitsma S. E., et al. (2018). New Insights in Type I and II CD20 Antibody Mechanisms-Of-Action with a Panel of Novel CD20 Antibodies. Br. J. Haematol. 180 (6), 808–820. 10.1111/bjh.15132 [DOI] [PubMed] [Google Scholar]
- Mondotte J. A., Lozach P. Y., Amara A., Gamarnik A. V. (2007). Essential Role of Dengue Virus Envelope Protein N Glycosylation at Asparagine-67 during Viral Propagation. J. Virol. 81 (13), 7136–7148. 10.1128/JVI.00116-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Scharf L., Euler Z., Liu Y., Eden C., Scheid J. F., et al. (2012). Complex-type N-Glycan Recognition by Potent Broadly Neutralizing HIV Antibodies. Proc. Natl. Acad. Sci. U S A. 109 (47), E3268–E3277. 10.1073/pnas.1217207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murin C. D., Julien J. P., Sok D., Stanfield R. L., Khayat R., Cupo A., et al. (2014). Structure of 2G12 Fab2 in Complex with Soluble and Fully Glycosylated HIV-1 Env by Negative-Stain Single-Particle Electron Microscopy. J. Virol. 88 (17), 10177–10188. 10.1128/JVI.01229-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Kawaoka Y. (2006). Host Range Restriction and Pathogenicity in the Context of Influenza Pandemic. Emerg. Infect. Dis. 12, 881–886. 10.3201/eid1206.051336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi R., Ohuchi M., Garten W., Klenk H. D. (1997). Oligosaccharides in the Stem Region Maintain the Influenza Virus Hemagglutinin in the Metastable Form Required for Fusion Activity. J. Virol. 71 (5), 3719–3725. 10.1128/JVI.71.5.3719-3725.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagadala N. S., Syed K., Tuszynski J. (2017). Software for Molecular Docking: a Review. Biophysical Rev. 9, 91. 10.1007/s12551-016-0247-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. J., Lee J., Qi Y., Kern N. R., Lee H. S., Jo S., et al. (2019). CHARMM-GUI Glycan Modeler for Modeling and Simulation of Carbohydrates and Glycoconjugates. Glycobiology 29 (4), 320–331. 10.1093/glycob/cwz003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R., Doores K. J., Walker L. M., Khayat R., Huang P. S., Wang S. K., et al. (2011). A Potent and Broad Neutralizing Antibody Recognizes and Penetrates the HIV Glycan Shield. Science 334 (6059), 1097–1103. 10.1126/science.1213256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R., Walker L. M., Stanfield R. L., Phogat S. K., Koff W. C., Poignard P., et al. (2010). Structure and Function of Broadly Reactive Antibody PG16 Reveal an H3 Subdomain that Mediates Potent Neutralization of HIV-1. Proc. Natl. Acad. Sci. U S A. 107, 11483. 10.1073/pnas.1004600107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipirou Z., Powlesland A. S., Steffen I., Pohlmann S., Taylor M. E., Drickamer K. (2011). Mouse LSECtin as a Model for a Human Ebola Virus Receptor. Glycobiology 21, 806–812. 10.1093/glycob/cwr008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PyMOL (2019). The PyMOL Molecular Graphics System, Version 2.3.4. New York: Schrödinger, LLC. [Google Scholar]
- Raman R., Tharakaraman K., Shriver Z., Jayaraman A., Sasisekharan V., Sasisekharan R. (2014). Glycan Receptor Specificity as a Useful Tool for Characterization and Surveillance of Influenza A Virus. Trends Microbiol. 22, 632. 10.1016/j.tim.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. E., Franco K., Elliott D. H., Maher M. J., Reyna A., Montefiori D. C., et al. (2010). Quaternary Epitope Specificities of Anti-HIV-1 Neutralizing Antibodies Generated in Rhesus Macaques Infected by the Simian/human Immunodeficiency Virus SHIVSF162P4. J. Virol. 84 (7), 3443–3453. 10.1128/JVI.02617-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollenske T., Szijarto V., Lukasiewicz J., Guachalla L. M., Stojkovic K., Hartl K., et al. (2018). Cross-specificity of Protective Human Antibodies against Klebsiella pneumoniae LPS O-Antigen. Nat. Immunol. 19 (6), 617–624. 10.1038/s41590-018-0106-2 [DOI] [PubMed] [Google Scholar]
- Sanders R. W., Derking R., Cupo A., Julien J. P., Yasmeen A., de Val N., et al. (2013). A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 Gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-neutralizing Antibodies. Plos Pathog. 9 (9), e1003618. 10.1371/journal.ppat.1003618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R. W., Venturi M., Schiffner L., Kalyanaraman R., Katinger H., Lloyd K. O., et al. (2002). The Mannose-dependent Epitope for Neutralizing Antibody 2G12 on Human Immunodeficiency Virus Type 1 Glycoprotein Gp120. J. Virol. 76 (14), 7293–7305. 10.1128/jvi.76.14.7293-7305.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan C. N., Pantophlet R., Wormald M. R., Ollmann Saphire E., Stanfield R., Wilson I. A., et al. (2002). The Broadly Neutralizing Anti-human Immunodeficiency Virus Type 1 Antibody 2G12 Recognizes a Cluster of Alpha1-->2 Mannose Residues on the Outer Face of Gp120. J. Virol. 76 (14), 7306–7321. 10.1128/jvi.76.14.7306-7321.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabright G. E., Cottrell C. A., van Gils M. J., D'addabbo A., Harvey D. J., Behrens A. J., et al. (2020). Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions. Structure 28 (8), 897–909.e6. 10.1016/j.str.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabright G. E., Doores K. J., Burton D. R., CrispinProtein M., Mimicry G. (2019). In HIV Vaccine Design. J. Mol. Biol. 431 (12), 2223–2247. 10.1016/j.jmb.2019.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirts M. R., Klein C., Swails J. M., Yin J., Gilson M. K., Mobley D. L., et al. (2017). Lessons Learned from Comparing Molecular Dynamics Engines on the SAMPL5 Dataset. J. Computer-Aided Mol. Des. 31, 147. 10.1007/s10822-016-9977-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. F., Cummings R. D. (2014). Investigating Virus-Glycan Interactions Using Glycan Microarrays. Curr. Opin. Virol. 7C, 79–87. 10.1016/j.coviro.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield R. L., De Castro C., Marzaioli A. M., Wilson I. A., Pantophlet R. (2015). Crystal Structure of the HIV Neutralizing Antibody 2G12 in Complex with a Bacterial Oligosaccharide Analog of Mammalian Oligomannose. Glycobiology 25 (4), 412–419. 10.1093/glycob/cwu123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stencel-Baerenwald J. E., Reiss K., Reiter D. M., Stehle T., Dermody T. S. (2014). The Sweet Spot: Defining Virus-Sialic Acid Interactions. Nat. Rev. Microbiol. 12, 739–749. 10.1038/nrmicro3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroh L. J., Stehle T. (2014). Glycan Engagement by Viruses: Receptor Switches and Specificity. Annu. Rev. Virol. 1, 285–306. 10.1146/annurev-virology-031413-085417 [DOI] [PubMed] [Google Scholar]
- Sun X., Jayaraman A., Maniprasad P., Raman R., Houser K. V., Pappas C., et al. (2013). N-Linked Glycosylation of the Hemagglutinin Protein Influences Virulence and Antigenicity of the 1918 Pandemic and Seasonal H1N1 Influenza A Viruses. J. Virol. 87 (15), 8756–8766. 10.1128/JVI.00593-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate M. D., Job E. R., Deng Y. M., Gunalan V., Maurer-Stroh S., Reading P. C. (2014). Playing Hide and Seek: How Glycosylation of the Influenza Virus Hemagglutinin Can Modulate the Immune Response to Infection. Viruses 6 (3), 1294–1316. 10.3390/v6031294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. E., Drickamer K. (2003). Introduction to Glycobiology. Oxford; New York: Oxford University Press. [Google Scholar]
- Tharakaraman K., Watanabe S., Chan K. R., Huan J., Subramanian V., Chionh Y. H., et al. (2018). Rational Engineering and Characterization of an mAb that Neutralizes Zika Virus by Targeting a Mutationally Constrained Quaternary Epitope. Cell Host Microbe 23 (5), 618–627.e6. 10.1016/j.chom.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., et al. (1996). Human Monoclonal Antibody 2G12 Defines a Distinctive Neutralization Epitope on the Gp120 Glycoprotein of Human Immunodeficiency Virus Type 1. J. Virol. 70 (2), 1100–1108. 10.1128/JVI.70.2.1100-1108.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breedam W., Pohlmann S., Favoreel H. W., de Groot R. J., Nauwynck H. J. (2014). Bitter-sweet Symphony: Glycan-Lectin Interactions in Virus Biology. FEMS Microbiol. Rev. 38, 598–632. 10.1111/1574-6976.12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Cummings R. D., Esko J. D., Freeze H. H., Hart G. W., Etzler M. E. (2008). Essentials of Glycobiology Edn Second Edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Wadstrom T., Ljungh A. (1999). Glycosaminoglycan-binding Microbial Proteins in Tissue Adhesion and Invasion: Key Events in Microbial Pathogenicity. J. Med. Microbiol. 48, 223–233. 10.1099/00222615-48-3-223 [DOI] [PubMed] [Google Scholar]
- Wagner R., Heuer D., Wolff T., Herwig A., Klenk H. D. (2002). N-Glycans Attached to the Stem Domain of Haemagglutinin Efficiently Regulate Influenza A Virus Replication. J. Gen. Virol. 83 (Pt 3), 601–609. 10.1099/0022-1317-83-3-601 [DOI] [PubMed] [Google Scholar]
- Walker L. M., Huber M., Doores K. J., Falkowska E., Pejchal R., Julien J. P., et al. (2011). Broad Neutralization Coverage of HIV by Multiple Highly Potent Antibodies. Nature 477 (7365), 466–470. 10.1038/nature10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Berndsen Z. T., Raghwani J., Seabright G. E., Allen J. D., Pybus O. G., et al. (2020). Vulnerabilities in Coronavirus Glycan Shields Despite Extensive Glycosylation. Nat. Commun. 11 (1), 2688. 10.1038/s41467-020-16567-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. J., Boyington J. C., Dai K., Houser K. V., Pearce M. B., Kong W. P., et al. (2010). Cross-neutralization of 1918 and 2009 Influenza Viruses: Role of Glycans in Viral Evolution and Vaccine Design. Sci. Transl Med. 2 (24), 24ra21. 10.1126/scitranslmed.3000799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesener D. A., Dugan A., Kiessling L. L. (2017). Recognition of Microbial Glycans by Soluble Human Lectins. Curr. Opin. Struct. Biol. 44, 168–178. 10.1016/j.sbi.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. P., Jr, Galimidi R. P., Foglesong C. P., Gnanapragasam P. N., Huey-Tubman K. E., Klein J. S., et al. (2009). Design and Expression of a Dimeric Form of Human Immunodeficiency Virus Type 1 Antibody 2G12 with Increased Neutralization Potency. J. Virol. 83 (1), 98–104. 10.1128/JVI.01564-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R. J., Tessier M. B. (2010). Computational Glycoscience: Characterizing the Spatial and Temporal Properties of Glycans and Glycan-Protein Complexes. Curr. Opin. Struct. Biol. 20 (5), 575–583. 10.1016/j.sbi.2010.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N. C., Thompson A. J., Xie J., Lin C. W., Nycholat C. M., Zhu X., et al. (2018). A Complex Epistatic Network Limits the Mutational Reversibility in the Influenza Hemagglutinin Receptor-Binding Site. Nat. Commun. 9 (1), 1264. 10.1038/s41467-018-03663-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., West A. P., Jr, Kim H. J., Thornton M. E., Ward A. B., Bjorkman P. J. (2013). Structural Basis for Enhanced HIV-1 Neutralization by a Dimeric Immunoglobulin G Form of the Glycan-Recognizing Antibody 2G12. Cell Rep 5 (5), 1443–1455. 10.1016/j.celrep.2013.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto F., Clausen H., White T., Marken J., Hakomori S. (1990). Molecular Genetic Basis of the Histo-Blood Group ABO System. Nature 345 (6272), 229–233. 10.1038/345229a0 [DOI] [PubMed] [Google Scholar]
- Yang M., Angles d'Ortoli T., Säwén E., Jana M., Widmalm G., MacKerell A. D. (2016). Delineating the Conformational Flexibility of Trisaccharides from NMR Spectroscopy Experiments and Computer Simulations. Phys. Chem. Chem. Phys. 18 (28), 18776–18794. 10.1039/c6cp02970a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Huang J., Simon R., Wang L. X., MacKerell A. D. (2017). Conformational Heterogeneity of the HIV Envelope Glycan Shield. Scientific Rep. 7, 4435. 10.1038/s41598-017-04532-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen S., Yang D., Wang X., Zhu J., Peng D., et al. (2015). Role of Stem Glycans Attached to Haemagglutinin in the Biological Characteristics of H5N1 Avian Influenza Virus. J. Gen. Virol. 96 (Pt 6), 1248–1257. 10.1099/vir.0.000082 [DOI] [PubMed] [Google Scholar]