Abstract
Background
Rapid tests to evaluate severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific T-cell responses are urgently needed to decipher protective immunity and aid monitoring vaccine-induced immunity.
Methods
Using a rapid whole blood assay requiring a minimal amount of blood, we measured qualitatively and quantitatively SARS-CoV-2-specific CD4 T-cell responses in 31 healthcare workers using flow cytometry.
Results
100% of COVID-19 convalescent participants displayed a detectable SARS-CoV-2-specific CD4 T-cell response. SARS-CoV-2-responding cells were also detected in 40.9% of participants with no COVID-19-associated symptoms or who tested PCR-negative. Phenotypic assessment indicated that, in COVID-19 convalescent participants, SARS-CoV-2 CD4 responses displayed an early differentiated memory phenotype with limited capacity to produce interferon (IFN)-γ. Conversely, in participants with no reported symptoms, SARS-CoV-2 CD4 responses were enriched in late differentiated cells, coexpressing IFN-γ and tumour necrosis factor-α and also Granzyme B.
Conclusions
This proof-of-concept study presents a scalable alternative to peripheral blood mononuclear cell-based assays to enumerate and phenotype SARS-CoV-2-responding T-cells, thus representing a practical tool to monitor adaptive immunity due to natural infection or vaccine trials.
Short abstract
This proof-of-concept study shows that SARS-CoV-2 T-cell responses are easily detectable using a rapid whole blood assay requiring minimal blood volume. Such assay represents a suitable tool to monitor adaptive immunity in vaccine trials. https://bit.ly/3yZHtcL
Introduction
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (causing the disease known as COVID-19), which first emerged in Wuhan, China, in December 2019, was declared a global pandemic on 12 March 2020 and is affecting all countries of the world, including those of Africa [1]. There is an urgent need to better understand the clinical manifestations and the pathogenesis of SARS-CoV-2 in order to develop relevant tools, including diagnostic tests, treatments and vaccines, to stop the spread of disease, as well as strategies to best manage this disease in all population groups.
The clinical spectrum of COVID-19 is very wide, from asymptomatic through mild flu-like symptoms to severe pneumonia and death. Understanding what constitutes immune protection against SARS-CoV-2 is key to predicting long-term immunity and to inform vaccine design. While much emphasis has been placed on B-cells and antibody response, it is not yet clear what type of immune response confers protection to SARS-CoV-2 [2]. Several studies suggest that the T-cell response may play an important role in SARS-CoV-2 pathogenesis and reports indicating that patients lacking B-cells can recover from SARS-CoV-2 infection further highlight the likely importance of T-cell immunity [3, 4]. Additionally, accumulating evidence indicates that the presence of pre-existing, cross-reactive memory T-cells specific for common cold coronaviruses may affect susceptibility to SARS-CoV-2 infection and partially explain the markedly divergent clinical manifestations of COVID-19 [5–8].
Efficient, sensitive and simple assessment of human T-cell immunity remains a challenge. Most commonly used T-cell assays necessitate the isolation of peripheral blood mononuclear cells (PBMCs), requiring a significant amount of blood. Therefore, whole blood assays could be more advantageous than PBMC-based methods by significantly reducing blood volume (∼1 mL), making them more applicable to paediatric populations. Moreover, such assays are rapid, as they do not require cell separation, and preserve the physiological cellular and soluble environments, better mimicking human blood conditions. Here, we report a rapid (∼7 h) whole blood-based detection method of SARS-CoV-2-specific T-cell responses with simple steps that could be adapted to settings with limited resources. This rapidly applicable assay could represent an easily standardisable tool to assess SARS-CoV-2-specific adaptive immunity to monitor T-cell responses in vaccine trials, gain insight into what constitutes a protective response or define the prevalence of SARS-CoV-2 T-cell responders population-wide.
Methods
Study population
The population studied consisted of healthcare workers (HCWs) (n=31, 29% male) recruited between July and September 2020 from Groote Schuur Hospital in Cape Town, the hardest hit region of the initial COVID-19 epidemic in South Africa [9]. Participants were classified according to 1) reporting of COVID-19-associated symptoms, 2) whether a SARS-CoV-2 RNA PCR from a nasal or pharyngeal swab was performed and 3) SARS-CoV-2 PCR results. Based on these criteria, participants were subdivided into three groups: 1) persons with no COVID-19-associated symptoms (n=15), 2) persons who reported symptoms but tested SARS-CoV-2 PCR-negative (n=7), and 3) persons who had COVID-19-associated symptoms and tested SARS-CoV-2 PCR-positive (n=9) (table 1). In all groups, the exposure to COVID-19 patients was comparable (86.8–100%). All participants with PCR-confirmed COVID-19 had mild symptoms and did not require hospitalisation. Blood samples were obtained a median of 7.3 weeks post-SARS-CoV-2 testing in persons with a negative test result and 4.7 weeks in those with a confirmed positive result (p=0.09). All participants were symptom-free at the time of sampling. In four SARS-CoV-2 PCR-positive participants, a second sample was obtained 1 month later. The University of Cape Town's Faculty of Health Sciences Human Research Ethics Committee approved the study (HREC 207/2020) and written informed consent was obtained from all participants.
TABLE 1.
Clinical characteristics of study participants (n=31 healthcare workers)
| p-value | ||||
| Participants (n) | 15 | 7 | 9 | NA |
| Self-reported COVID-19-like symptoms | No | Yes | Yes | NA |
| Tested for COVID-19 (PCR) | No | Yes | Yes | NA |
| Positive SARS-CoV-2 PCR | NA | No | Yes | NA |
| Positive SARS-CoV-2 N-specific IgG (%)# | 0 | 0 | 88.9 | NA |
| Presence of SARS-CoV-2 nAbs (%)¶ | 0 | 0 | 100 | NA |
| Contact with COVID-19 patients (%) | 86.6 | 100 | 88.9 | ns |
| Median (IQR) time post-PCR test (weeks) | NA | 7.3 (6.3–8.4) | 4.7 (2.6–7.2) | 0.09 |
NA: not applicable; N: nucleocapsid; nAb: neutralising antibody; IQR: interquartile range. #: measured using the Elecsys system; ¶: measured using a SARS-CoV-2 pseudovirus neutralisation assay. ns: nonsignificant.
Measurement of SARS-CoV-2 nucleocapsid-specific IgG in plasma
The measurement of SARS-CoV-2-specific antibodies was performed using the Elecsys Anti-SARS-CoV-2 immunoassay (Roche Diagnostics, Basel, Switzerland). This semiquantitative electrochemiluminescence immunoassay measures SARS-CoV-2 nucleocapsid-specific IgG. The assay was performed by the South African National Health Laboratory Service and interpreted according to the manufacturer's instructions (Roche: version 1.0 2020-05). Results are reported as numeric values in the form of a cut-off index (COI) (signal sample/cut-off), where a COI <1.0 corresponds to nonreactive plasma and a COI ⩾1.0 corresponds to reactive plasma. At 14 days post-SARS-CoV-2 PCR confirmation, the sensitivity and specificity of the Elecsys Anti-SARS-CoV-2 immunoassay has been reported as 99.5% (95% CI 97.0–100.0%) and 99.80% (95% CI 99.69–99.88%), respectively [10–12].
SARS-CoV-2 pseudovirus neutralisation assay
Patient plasma was evaluated for SARS-CoV-2 neutralisation activity using a SARS-CoV-2 pseudovirus infection assay. Single-cycle infectious SARS-CoV-2 pseudovirions based on the HIV backbone expressing the SARS-CoV-2 spike protein and a firefly luciferase reporter were produced in HEK-293TT cells [13, 14] by cotransfection of plasmids pNL4-3.Luc.R-E- (ARP-3418; NIH AIDS Reagent Program, Germantown, MD, USA) and pcDNA3.3-SARS-CoV-2-spike Δ18 [15]. HEK-293TT cell culture supernatants containing the virions were harvested 3 days post-transfection and incubated with heat-inactivated patient plasma at 5-fold serial dilutions for 60 min at 37°C. Plasma/pseudovirus mixtures were then used for transfection of HEK-293T cells stably expressing the angiotensin-converting enzyme 2 receptor [16]. Cells were lysed 3 days post-infection using the Promega cell culture lysis reagent (Promega Biosciences, San Luis Obispo, CA, USA) and assessed for luciferase activity using a GloMax Explorer Multimode Microplate Reader (Promega Biosciences) together with the Luciferase assay system (Promega Biosciences).
Whole blood-based T-cell detection assay
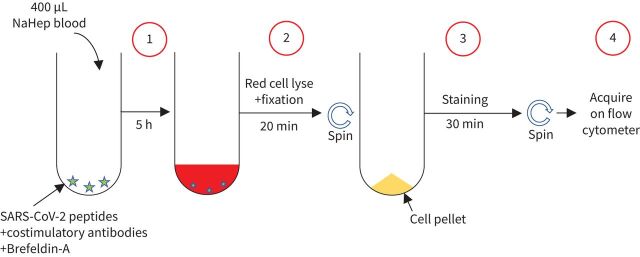
Blood was collected in sodium heparin tubes and processed within 3 h of collection. The whole blood-based SARS-CoV-2-specific T-cell detection assay was adapted from a previously reported whole blood intracellular cytokine detection assay designed to quantitate Mycobacterium tuberculosis-specific T-cells in small volumes of blood [17]. However, significant modifications have been made, including a reduced incubation time and the use of a fixation buffer allowing the simultaneous lysis of red blood cells to streamline processing time, leading to faster acquisition of results [18]. Here, we adapted this assay to detect SARS-CoV-2-specific T-cells using synthetic SARS-CoV-2 PepTivator peptides (Miltenyi Biotec, Woking, UK), consisting of 15mer sequences with 11-amino-acid overlap covering the immunodominant parts of the spike protein, and the complete sequence of the nucleocapsid and membrane proteins. All peptides were combined in a single pool and used at a final concentration of 1 µg·mL−1. The workflow of the assay is presented in figure 1. Briefly, 400 µL whole blood was stimulated with the SARS-CoV-2 spike, nucleocapsid and membrane protein peptide pool at 37°C for 5 h in the presence of costimulatory antibodies against CD28 and CD49d (1 µg·mL−1 each; BD Biosciences, San Jose, CA, USA) and Brefeldin-A (10 µg·mL−1; Sigma-Aldrich, St Louis, MO, USA). Unstimulated blood was incubated with costimulatory antibodies, Brefeldin-A and an equimolar amount of dimethyl sulfoxide (DMSO). Red blood cell lysis and white cell fixation was then performed as a single step using a transcription factor fixation buffer (eBioscience, San Diego, CA, USA) for 20 min. At this stage cells were cryopreserved in freezing media (50% fetal bovine serum, 40% RPMI and 10% DMSO) and stored in liquid nitrogen until batched analysis.
FIGURE 1.
Schematic showing methodology and workflow of the whole blood assay for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific adaptive immune responses. Step 1: 400 µL heparinised (NaHep) whole blood is incubated for 5 h in the presence of a SARS-CoV-2-specific peptide pool in the presence of costimulatory antibodies (i.e. CD28 and CD49d) and Brefeldin-A. Step 2: cells are incubated for 20 min in the presence of a transcription factor fixation buffer, leading to the simultaneous lysis of red blood cells and cell fixation. Step 3: cells are stained for 30 min with an optimised panel of fluorophore-labelled antibodies. Step 4: samples are acquired on a flow cytometer. Control samples are processed with a similar workflow in the absence of the SARS-CoV-2-specific peptide pool.
Cell staining was performed on cryopreserved cells that were thawed, washed and permeabilised with a transcription factor perm/wash buffer (eBioscience). Cells were then stained at room temperature for 30 min with antibodies for CD3 BV650, CD4 BV785, CD8 BV510, CD45RA Alexa 488, CD27 PE-Cy5, CD38 APC, HLA-DR BV605, Ki-67 PerCP-Cy5.5, PD-1 PE, Granzyme B (GrB) BV421, interferon (IFN)-γ BV711, tumour necrosis factor (TNF)-α PE-Cy7 and interleukin (IL)-2 PE/Dazzle 594, as detailed in supplementary table S1. Samples were acquired on a BD LSR II flow cytometer and analysed using FlowJo version 9.9.6 (FlowJo, Ashland, OR, USA). A positive response was defined as any cytokine response that was at least twice the background of unstimulated cells. To define the phenotype of SARS-CoV-2-specific CD4 T-cells, a cut-off of 30 events was used. The gating strategy is provided in supplementary figure S1.
Statistical analyses
Graphical representations were performed in Prism version 8.4.3 (GraphPad, San Diego, CA, USA) and JMP version 14.0.0 (SAS Institute, Cary, NC, USA). Statistical tests were performed in Prism. Nonparametric tests were used for all comparisons. The Kruskal–Wallis test with Dunn's multiple comparison test was used for multiple comparisons, and the Mann–Whitney and Wilcoxon matched pairs test was used for unmatched and paired samples, respectively.
Results
Serological assessment of SARS-CoV-2 sensitisation
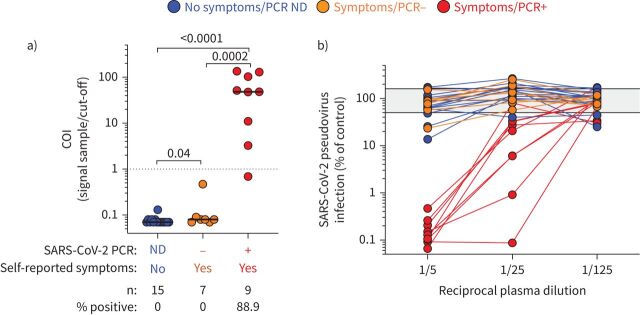
Even though real-time reverse transcription-PCR is the most specific technique to detect acute SARS-CoV-2 infection, the positivity rate drops rapidly as soon as 10 days post-symptom onset, particularly in individuals with mild forms of COVID-19 [19, 20]. Hence, serology assays provide an important complement to RNA testing to identify individuals who have been sensitised by SARS-CoV-2. Thus, to assess potential SARS-CoV-2 sensitisation in participants who did not have a SARS-CoV-2 PCR test performed or tested PCR-negative, the presence of SARS-CoV-2-specific antibodies (e.g. SARS-CoV-2 nucleocapsid-specific IgG) was measured and a SARS-CoV-2 pseudovirus neutralisation assay was also performed in all participants. While all participants with a positive SARS-CoV-2 PCR test exhibited in vitro anti-SARS-CoV-2 neutralising activity and eight out of nine (88.8%) had detectable SARS-CoV-2 nucleocapsid-specific IgG, none of the participants who tested negative or did not undergo PCR testing were positive for SARS-CoV-2 nucleocapsid-specific IgG (figure 2a) or displayed robust in vitro anti-SARS-CoV-2 activity (figure 2b). Overall, these results confirmed that PCR-positive participants had been infected with SARS-CoV-2 and mounted an immune response to the virus. For the other participants, it is difficult to ascertain their SARS-CoV-2 infection status. On the one hand, the absence of antibody response and neutralisation activity may suggest that they have not been SARS-CoV-2-infected. On the other hand, due to their high level of SARS-CoV-2 exposure, we cannot fully exclude SARS-CoV-2 infection, as while uncommon, COVID-19-confirmed patients with negative antibody results have been reported [21].
FIGURE 2.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) serological assessment. a) Quantification of SARS-CoV-2 nucleocapsid-specific antibodies using the Elecsys assay expressed as a cut-off index (COI) (signal sample/cut-off). Participants were grouped according to their clinical characteristics. Medians (black bar) are shown. The dotted line indicates the manufacturer's cut-off value for positivity. Statistical comparisons were performed using the Mann–Whitney t-test; p-values are shown. b) SARS-CoV-2 pseudovirus neutralisation activity. SARS-CoV-2 pseudovirions pre-incubated with serially diluted patient plasma were used to infect angiotensin-converting enzyme 2-expressing HEK-293T cells. Luciferase activity as a measure for infection was assessed 3 days post-infection and results are expressed as infection compared with control (untreated virions, grey shaded area), which was set at 100%. ND: not done.
Magnitude and functional profile of SARS-CoV-2-responding CD4 T-cells
Among the 31 participants tested, 58% (n=18) had a detectable SARS-CoV-2-specific CD4 T-cell response (producing any of the measured cytokines: IFN-γ, TNF-α or IL-2) using the described whole blood assay (figure 3a). We then defined the magnitude and phenotype of SARS-CoV-2-specific CD4 responses according to participants’ clinical characteristics (figure 3b and c). All HCWs with self-reported symptoms and a SARS-CoV-2 PCR-positive test (n=9) were found to have SARS-CoV-2-specific CD4 T-cells (median (IQR) frequency 0.4% (0.28–0.65%)) (figure 3c). Of note, the frequency of SARS-CoV-2-specific CD4 T-cells did not associate with the magnitude of SARS-CoV-2 nucleocapsid-specific IgG (p=0.64, r= −0.18) or plasma neutralising activity (p=0.46, r=0.28) (data not shown). Interestingly, in participants with no detectable SARS-CoV-2 nucleocapsid-specific IgG or neutralising antibody activity, of those with self-reported symptoms, three out of seven (43%) had detectable SARS-CoV-2-responding CD4 T-cells (median (IQR) of responders 0.02% (0.019–0.03%)) and six out of 15 (40%) of those with no self-reported symptoms had detectable SARS-CoV-2-responding CD4 T-cells (median (IQR) of responders 0.15% (0.05–1.47%)). Moreover, a SARS-CoV-2-specific CD8 T-cell response was also observed in six out of the nine SARS-CoV-2 PCR-positive participants (66.7%). In participants reporting no symptoms (n=15), only one person had a detectable CD8 response and none of the PCR-negative participants (n=7) exhibited a detectable SARS-CoV-2-specific CD8 T-cell response (data not shown).
FIGURE 3.
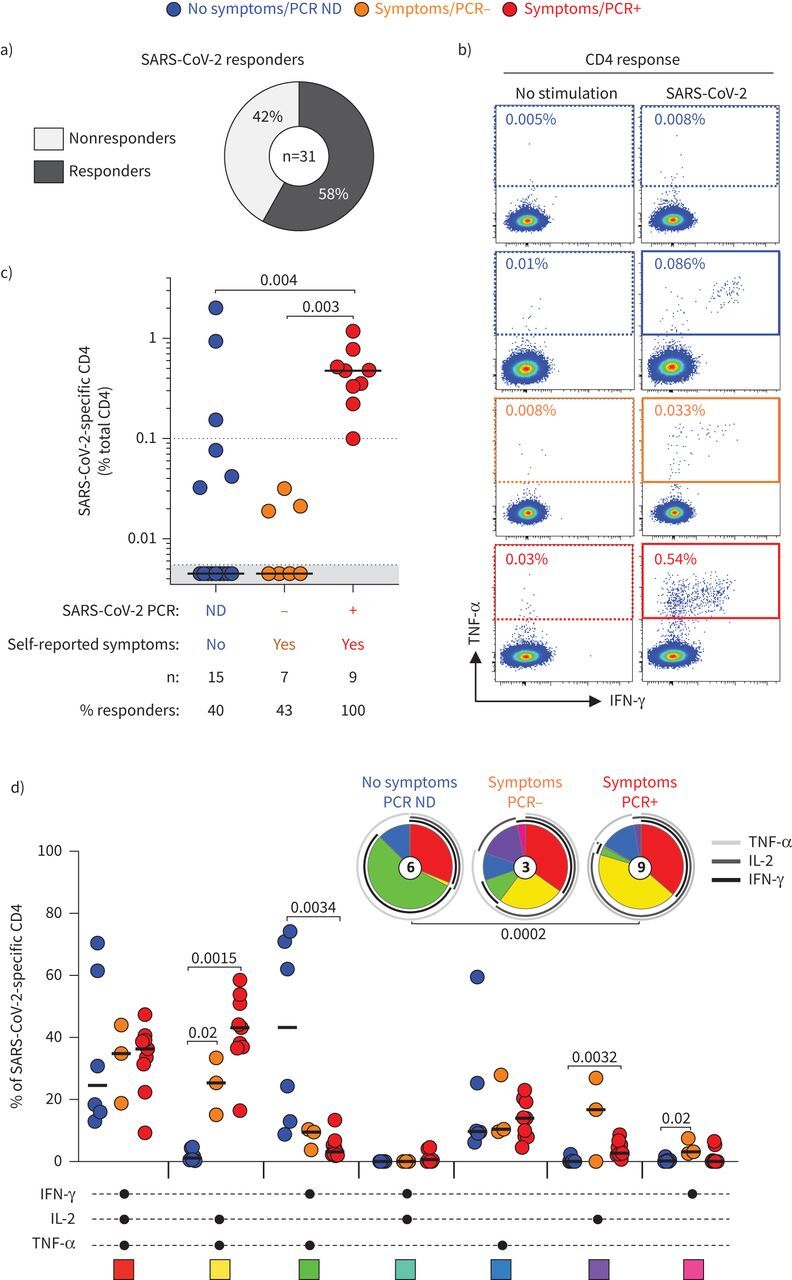
Magnitude and functional profile of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific CD4 T-cells. a) Proportion of participants exhibiting a detectable SARS-CoV-2-specific CD4 T-cell response. b) Representative examples of tumour necrosis factor (TNF)-α and interferon (IFN)-γ production in CD4 T-cells in response to the SARS-CoV-2 peptide pool. c) Magnitude of SARS-CoV-2-specific CD4 T-cell response (expressed as a percentage of total CD4 T-cells) in participants grouped according to their clinical characteristics. The number of participants and percentage of responders in each group is presented at the bottom of the graph. Statistical comparisons were performed using the Kruskal–Wallis test; p-values are shown. d) Polyfunctional profile of SARS-CoV-2-specific CD4 T-cells in each group. The x-axis displays the composition of each combination which is denoted with a black circle for the presence of IFN-γ, interleukin (IL)-2 and TNF-α. Medians (black bar) are shown. Each combination is colour-coded and data are summarised in the pie charts, where each pie slice represents the median contribution of each combination to the total SARS-CoV-2 response. The arcs identify the contribution of TNF-α, IL-2 and IFN-γ to the SARS-CoV-2 response. The Wilcoxon rank sum test was used to compare response patterns between groups and statistical differences between pie charts were defined using the permutation test; p-values are shown. ND: not done.
Next, we defined the polyfunctional profile of SARS-CoV-2-responding CD4 T-cells based on their capacity to coexpress IL-2, IFN-γ or TNF-α (figure 3d). The overall functional profile of SARS-CoV-2-specific cells in PCR-positive participants was distinct from participants with no reported symptoms (p=0.0002). Indeed, the SARS-CoV-2-specific CD4 response in PCR-positive participants was characterised by limited expression of IFN-γ, and was enriched in cells coexpressing IL-2 and TNF-α. On the contrary, in participants reporting no symptoms, most SARS-responding CD4 cells were distributed between triple functional cells (IL-2+IFN-γ+TNF-α+) and cells coproducing IFN-γ and TNF-α. Overall, in this assay, TNF-α was the predominant cytokine produced, with its production being significantly higher compared with IL-2 (p=0.0026) and IFN-γ (p<0.0001) (supplementary figure S2).
Phenotypic assessment of SARS-CoV-2-responding CD4 T-cells
While the proposed assay can be performed using a limited antibody panel to identify the frequency of SARS-CoV-2-responding T-cells, by solely measuring cytokine production, the use of a more extensive antibody panel also permits definition of the phenotypic profile of these cells. To this end, we included additional markers to assess the memory differentiation (CD27 and CD45RA), cytotoxic potential (GrB) and activation profile (CD38, HLA-DR, Ki-67 or PD-1) of SARS-CoV-2-responding CD4 T-cells.
Figure 4a and b shows that in all individuals with symptoms, SARS-CoV-2-specific CD4 T-cells displayed almost exclusively (median 97.7%) an early differentiated memory phenotype (ED: CD45RA−CD27+). On the contrary, in four out of six HCWs with no symptoms, SARS-CoV-2-specific CD4 T-cells exhibited a predominant late differentiated phenotype (LD: CD45RA−CD27−).
FIGURE 4.
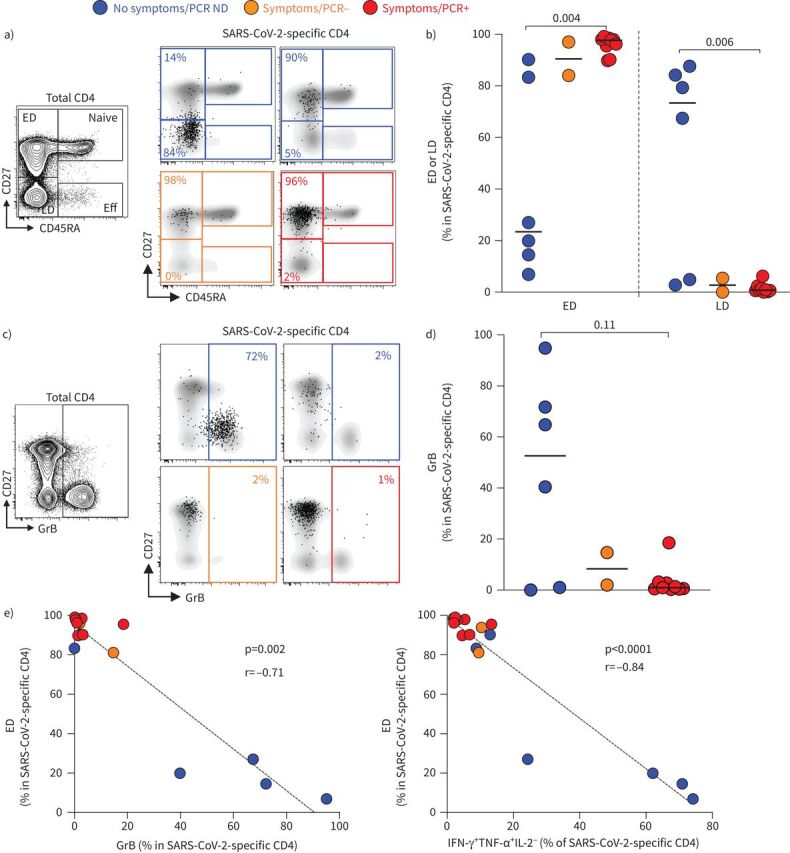
Memory differentiation profile and Granzyme B (GrB) expression in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific CD4 T-cells. a) Representative examples of the memory differentiation profile of SARS-CoV-2-specific CD4 T-cells based on the expression of CD45RA and CD27. The flow plot on the right shows the distribution of naive (CD45RA+CD27+), early differentiated (ED: CD45RA−CD27+), late differentiated (LD: CD45RA−CD27−) and effector (Eff: CD45RA+CD27−) cells in total CD4 T-cells. b) Summary graph of the proportion of ED and LD in SARS-CoV-2-specific CD4 T-cells in each group. Statistical comparisons were performed using the Kruskal–Wallis test; p-values are shown. c) Representative examples of GrB expression in total and SARS-CoV-2-specific CD4 T-cells. d) Summary graph of GrB expression in SARS-CoV-2-specific CD4 T-cells in each group. Statistical comparisons were performed using the Kruskal–Wallis test; p-value is shown. e) Relationship between the proportion of ED within SARS-CoV-2-specific CD4 T-cells and GrB expression or the proportion of IFN-γ+TNF-α+IL-2− SARS-CoV-2-specific CD4 T-cells. Correlations were tested by the two-tailed nonparametric Spearman rank test. ND: not done; IFN: interferon; TNF: tumour necrosis factor; IL: interleukin.
While the role of cytotoxic CD4 T-cells is still unclear, the presence of these cells has been described in several viral infections [22]. We thus measured GrB expression in SARS-CoV-2-responding CD4 T-cells. While GrB was barely detectable in SARS-CoV-2-specific CD4 T-cells in PCR-positive participants, elevated GrB expression was observed in four out of six participants without symptoms (figure 4c and d). Moreover, the proportion of ED SARS-CoV-2-responding CD4 T-cells inversely associated with GrB expression (p=0.002, r= −0.71) and the proportion of IFN-γ and TNF-α dual-producing cells (p<0.0001, r= −0.84) (figure 4e). The phenotypic characteristics of GrB-expressing SARS-CoV-2-responding CD4 T-cells (i.e. highly differentiated, and producing IFN-γ and TNF-α) are in agreement with a recent report describing cytomegalovirus (CMV)-specific CD4 cytotoxic T-lymphocyte T-cells [23].
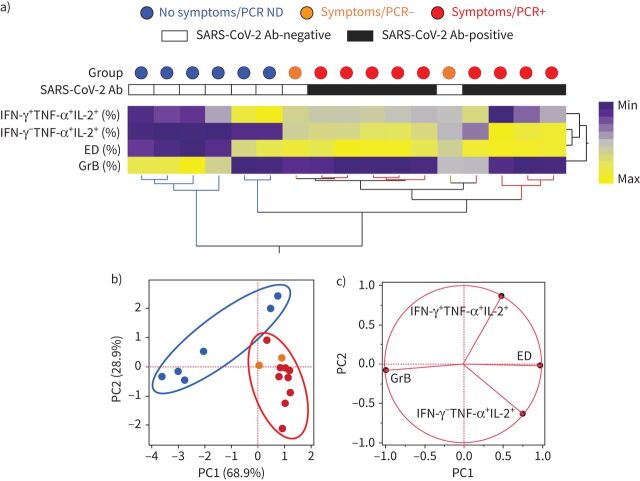
To determine if the overall phenotypic profile of SARS-CoV-2-responding CD4 T-cells allowed discrimination of participants based on their clinical characteristics, we performed a hierarchical clustering analysis (figure 5a) and a principal component analysis (figure 5b), including four parameters (i.e. proportions of IFN-γ+TNF-α+IL-2+ and IFN-γ−TNF-α+IL-2+ cells, proportion of ED, and GrB expression). Both analyses show that participants who reported no symptoms and were negative for SARS-CoV-2 antibodies separated clearly from PCR-positive participants. Conversely, PCR and SARS-CoV-2 antibody-negative HCWs reporting symptoms could not be separated from PCR-positive participants. The loading plot shows that GrB expression and the proportion of triple-positive cells were the main drivers permitting the segregation of participants who reported no symptoms and were negative for SARS-CoV-2 antibodies (figure 5c)
FIGURE 5.
Phenotypic signature of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific IFN-γ+ CD4+ T-cells according to clinical characteristics. a) Nonsupervised two-way hierarchical cluster analysis (Ward method) using four phenotypic parameters (i.e. proportions of IFN-γ+TNF-α+IL-2+ and IFN-γ−TNF-α+IL-2+ cells, proportion of ED, and GrB expression) from SARS-CoV-2-specific CD4 T-cells. Each column represents a participant and is colour-coded according to their clinical characteristics indicated by a circle at the top of the dendrogram. Participants with a positive or negative SARS-CoV-2 serology test are indicated. Data are depicted as a heatmap coloured from minimum to maximum values for each parameter. b) Principal component analysis on correlations, derived from the three studied parameters. Each data point represents a participant. The two axes represent principal components 1 (PC1) and 2 (PC2). Their contribution to the total data variance is shown as a percentage. c) Loading plot showing how each parameter influences PC1 and PC2 values. ND: not done; Ab: antibody; IFN: interferon; TNF: tumour necrosis factor; IL: interleukin.
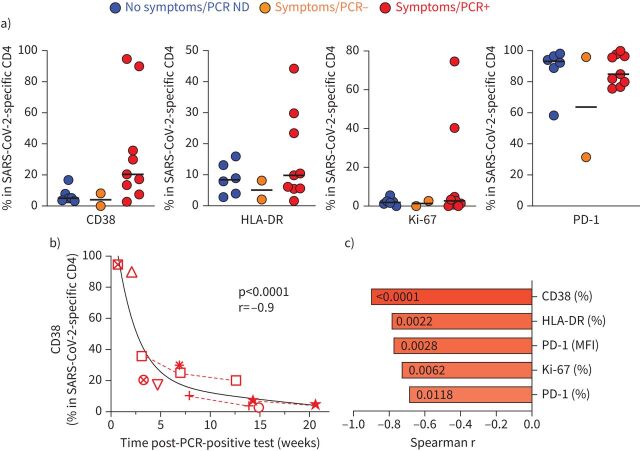
Lastly, as the expression of activation markers (CD38, HLA-DR, Ki-67 or PD-1) on antigen-specific T-cells is indicative of active infection [24, 25], we defined the expression of these markers on SARS-CoV-2-responding CD4 T-cells. No significant difference was observed in any of the measured markers among the different groups, notwithstanding a few outliers observed in the PCR-positive participant group (figure 6a). Interestingly, regardless of the participants’ clinical characteristics, PD-1 was highly expressed on SARS-CoV-2-responding CD4 T-cells. We then defined the relationship between the activation profile of SARS-CoV-2-specific CD4 T-cells and time post-PCR-positive test. Figure 6b shows that in PCR-positive participants, the expression of CD38 associated strongly with the time post-PCR testing (p<0.0001, r= −0.91), where CD38 expression decreased sharply in the first 2–3 weeks post-testing, suggesting a rapid clearance of the pathogen. Similar associations were observed for all tested activation markers, with CD38 expression showing the strongest correlation (figure 6c).
FIGURE 6.
Activation profile of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific CD4 T-cells. a) CD38, HLA-DR, Ki-67 and PD-1 expression in SARS-CoV-2-specific CD4 T-cells in each group. Medians (black bar) are shown. No significant differences were observed between groups for any markers using the Kruskal–Wallis test. b) Association between CD38 expression in SARS-CoV-2-specific CD4 T-cells and the time post-SARS-CoV-2 PCR-positive test. Each symbol represents a participant (n=9). Dashed red lines identify participants with longitudinal samples. Correlations were tested by the two-tailed nonparametric Spearman rank test. c) Comparison of the correlation between the time post-SARS-CoV-2 PCR-positive test and the expression of different activation profile markers, ranked according to the strength of the association. Spearman correlation r-values are plotted on the x-axis and corresponding p-values are shown within each bar. ND: not done; MFI: mean fluorescence intensity.
Discussion
In this proof-of-concept analysis assessing the use of a simple whole blood assay to measure SARS-CoV-2-specific T-cell responses in a small HCW cohort, we show that SARS-CoV-2-specific CD4 T-cells (expressing IFN-γ, TNF-α or IL-2) were easily detectable in all tested convalescent COVID-19 participants. However, in participants who did not experience any COVID-19-related symptoms or tested SARS-CoV-2 PCR-negative (despite reporting symptoms), nine out of 22 (40.9%) also exhibited a detectable SARS-CoV-2-specific CD4 T-cell response. Nevertheless, the median frequency of SARS-CoV-2-specific CD4 T-cells in the latter groups was ∼5-fold lower compared with SARS-CoV-2 PCR-positive participants. Importantly, unlike convalescent COVID-19 participants, none of participants without symptoms or who tested PCR-negative had detectable SARS-CoV-2 nucleocapsid-specific IgG or displayed robust neutralising activity. Several hypotheses could explain the detection of SARS-CoV-2-responding cells in the absence of detectable antibody response. As the participants included in this study were most likely highly exposed to SARS-CoV-2 due to their occupation, one could argue that any SARS-CoV-2 CD4 T-cell response was 1) generated during an asymptomatic COVID-19 episode that did not result in a detectable antibody response, 2) reflecting recent exposure to SARS-CoV-2 or 3) corresponding to SARS-CoV-2 cross-reactive memory CD4 T-cells. A number of studies have demonstrated the presence of SARS-CoV-2-reactive CD4 T-cells in 40–60% of SARS-CoV-2-unexposed individuals in different populations around the world [26–29]. Additionally, it is important to point out that all these studies were performed using PBMCs and it is encouraging to find that a whole blood-based assay using a limited amount of blood (<1 mL) yields comparable results.
Further analyses of the polyfunctional and phenotypic profile of SARS-CoV-2-specific CD4 T-cells revealed that regardless of the clinical characteristics of the participants, the most prevalent cytokine detected in response to SARS-CoV-2 peptides was TNF-α. These observations are in accordance with other studies pointing out an impairment of SARS-CoV-2 adaptive immune responses characterised by low levels of type I and type II IFN [30, 31]. However, it is also possible that the limited IFN-γ production we observed in this study is related to the short stimulation time (e.g. 5 h) used in the assay. Nevertheless, this suggests that TNF-α could be a more reliable target than IFN-γ to detect SARS-CoV-2-specific CD4 T-cell responses. Additionally, SARS-CoV-2-responding CD4 T-cells in all participants were characterised by elevated PD-1 expression. Such profiles have been observed in acute SARS-CoV-2 infection and are likely driven by ongoing viral replication [27, 32, 33]. Expression of PD-1 on antigen-specific memory T-cells in the absence of acute infection could be surprising. However, depending on their specificity, antigen-specific memory T-cells can display a highly variable differentiation profile [34] and the maintenance of elevated PD-1 expression on some virus-specific memory T-cells (such as CMV or hepatitis C virus) has been reported even after infection clearance [35–37]. Moreover, a recent publication showed that PD-1-expressing SARS-CoV-2-specific CD8 T-cells are not exhausted but functional in convalescent COVID-19 patients [38]. Further experiments will be needed to understand the impact of elevated PD-1 expression of SARS-CoV-2-responding CD4 T-cells.
Lastly, SARS-CoV-2-responding CD4 T-cells were qualitatively different between convalescent COVID-19 participants and participants who did not experience COVID-19-associated symptoms. In the former group, SARS-CoV-2-specific CD4 T-cells displayed almost exclusively an ED memory phenotype; in the latter group, SARS-CoV-2-responsive CD4 T-cells exhibited a LD memory phenotype and were enriched in GrB.
Our study has several limitations. Our results rely on a small number of participants and need to be confirmed in larger cohorts. In the absence of a low SARS-CoV-2 exposure control group, we cannot speculate about the exact specificity of SARS-CoV-2-responding CD4 T-cells detected in participants who are serology-negative.
Moreover, technical optimisation and validation are warranted to streamline the proposed assay and test its performance. A comparable approach using a whole blood IFN-γ release assay (IGRA), commonly used for the detection of M. tuberculosis infection, has been recently applied to detect SARS-CoV-2-specific T-cell response [39]. While both techniques have a comparable cost (∼USD 30–40), the flow cytometry-based assay proposed in this study has a lower turnaround time compared with IGRA (∼7 versus 18 h), but more importantly our approach is more flexible. It can be used to simply quantify SARS-CoV-2-responding CD4 and CD8 T-cells (using a minimal four-colour panel) to monitor the frequency of T-cell responses in epidemiological studies or vaccine trials, or be used for more exploratory work assessing in-depth the phenotype and functionality of SARS-CoV-2-specific T-cells. Regarding the sensitivity and specificity of each assay, parallel analyses will need to be performed to compare the performance of both assays. Additionally, further optimisations should be performed to improve the standardisation and flexibility of the proposed SARS-CoV-2-specific whole blood assay, including using tubes containing lyophilised reagents for cell stimulation, pre-mixed antibody cocktail for cell staining and automated gating for flow analyses. Another alternative to alleviate collection time and transport constraints, and for limited-resource sites, would be to directly cryopreserve whole blood samples using cryopreservative solution, such as CryoStor CS10 solution [40], allowing the cell stimulation and staining to be performed in batch at adequately equipped sites. Lastly, in this study, cell stimulation was performed using a pool of peptides covering the SARS-CoV-2 spike, nucleocapsid and membrane proteins. In light of a recent publication showing that the discrimination between a pre-existing cross-reactive response and COVID-19-induced response is dependent on the specificity of the SARS-CoV-2 antigen used [41], it appears that the peptide combination used in this study is not optimal to distinguished pre-existing cross-reactive SARS-CoV-2-reactive T-cells from COVID-19-induced responses. Thus, the proposed assay may be further enhanced by using more targeted peptide pools with defined specificity for SARS-CoV-2 or seasonal coronaviruses.
Nevertheless, the primary objective of this study was to describe, as a proof of concept, a rapid and flexible whole blood assay, requiring a very limited amount of blood, that could represent a powerful tool to investigate SARS-CoV-2-specific T-cell responses both quantitatively and qualitatively even in COVID-19 paediatric cases.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00285-2021.SUPPLEMENT (479.9KB, pdf)
Shareable PDF
Acknowledgements
The authors thank all the participants.
Footnotes
This article has supplementary material available from erj.ersjournals.com
Author contributions: C. Riou, K.A. Wilkinson and R.J. Wilkinson designed the study. C. Riou performed the whole blood assay and flow cytometry experiments, and analysed and interpreted the data. G. Schäfer performed the neutralising assay experiments. E. du Bruyn, R.T. Goliath and C. Stek recruited the study participants. K.A. Wilkinson, H. Mou and D. Hung provided critical reagents. C. Riou and K.A. Wilkinson wrote the manuscript, with all authors contributing to providing critical feedback.
Conflict of interest: C. Riou has nothing to disclose.
Conflict of interest: G. Schäfer has nothing to disclose.
Conflict of interest: E. du Bruyn has nothing to disclose.
Conflict of interest: R.T. Goliath has nothing to disclose.
Conflict of interest: C. Stek has nothing to disclose.
Conflict of interest: H. Mou has nothing to disclose.
Conflict of interest: D. Hung has nothing to disclose.
Conflict of interest: K.A. Wilkinson has nothing to disclose.
Conflict of interest: R.J. Wilkinson reports grants from Wellcome, Cancer Research UK, UK Research and Innovation, and the European and Developing Countries Clinical Trials Partnership, during the conduct of the study.
Support statement: This work was supported by Wellcome (104803 and 203135) and the Crick Idea to Innovation (i2i) scheme in partnership with the Rosetrees Trust (2020–0009). C. Riou is supported by the European and Developing Countries Clinical Trials Partnership EDCTP2 programme supported by the European Union's Horizon 2020 programme (Training and Mobility Action TMA2017SF-1951-TB-SPEC) and the US National Institutes of Health (NIH) (R21AI148027). G. Schäfer is supported by the European and Developing Countries Clinical Trials Partnership EDCTP2 programme (Training and Mobility Action TMA2018SF-2446). R.J. Wilkinson and K.A. Wilkinson receive support from the Francis Crick Institute, which is funded by the UK Research and Innovation Medical Research Council, Cancer Research UK and Wellcome (FC0010218). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Margolin E, Burgers WA, Sturrock ED, et al. . Prospects for SARS-CoV-2 diagnostics, therapeutics and vaccines in Africa. Nat Rev Microbiol 2020; 18: 690–704. doi: 10.1038/s41579-020-00441-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox RJ, Brokstad KA. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat Rev Immunol 2020; 20: 581–582. doi: 10.1038/s41577-020-00436-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altmann DM, Boyton RJ. SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci Immunol 2020; 5: eabd6160. doi: 10.1126/sciimmunol.abd6160 [DOI] [PubMed] [Google Scholar]
- 4.Quinti I, Lougaris V, Milito C, et al. . A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol 2020; 146: 211–213. doi: 10.1016/j.jaci.2020.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canete PF, Vinuesa CG. COVID-19 makes B cells forget, but T cells remember. Cell 2020; 183: 13–15. doi: 10.1016/j.cell.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sette A, Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol 2020; 20: 457–458. doi: 10.1038/s41577-020-0389-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipsitch M, Grad YH, Sette A, et al. . Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol 2020; 20: 709–713. doi: 10.1038/s41577-020-00460-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun J, Loyal L, Frentsch M, et al. . SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020; 587: 270–274. doi: 10.1038/s41586-020-2598-9 [DOI] [PubMed] [Google Scholar]
- 9.Mendelson M, Boloko L, Boutall A, et al. . Clinical management of COVID-19: experiences of the COVID-19 epidemic from Groote Schuur Hospital, Cape Town, South Africa. S Afr Med J 2020; 110: 973–981. doi: 10.7196/SAMJ.2020.v110i10.15157 [DOI] [PubMed] [Google Scholar]
- 10.Muench P, Jochum S, Wenderoth V, et al. . Development and validation of the Elecsys anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J Clin Microbiol 2020; 58: e01694-20. doi: 10.1128/JCM.01694-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favresse J, Eucher C, Elsen M, et al. . Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS-CoV-2 total antibodies. Clin Chem 2020; 66: 1104–1106. doi: 10.1093/clinchem/hvaa131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National SARS-CoV-2 Serology Assay Evaluation Group . Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis 2020; 20: 1390–1400. doi: 10.1016/S1473-3099(20)30634-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buck CB, Pastrana DV, Lowy DR, et al. . Efficient intracellular assembly of papillomaviral vectors. J Virol 2004: 78; 751–757. doi: 10.1128/JVI.78.2.751-757.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buck CB, Pastrana DV, Lowy DR, et al. . Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 2005; 119: 445–462. doi: 10.1385/1-59259-982-6:445 [DOI] [PubMed] [Google Scholar]
- 15.Rogers TF, Zhao F, Huang D, et al. . Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020; 369: 956–963. doi: 10.1126/science.abc7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mou H, Quinlan BD, Peng H, et al. . Mutations from bat ACE2 orthologs markedly enhance ACE2-Fc neutralization of SARS-CoV-2. bioRxiv 2020; preprint [ 10.1101/2020.06.29.178459]. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanekom WA, Hughes J, Mavinkurve M, et al. . Novel application of a whole blood intracellular cytokine detection assay to quantitate specific T-cell frequency in field studies. J Immunol Methods 2004; 291: 185–195. doi: 10.1016/j.jim.2004.06.010 [DOI] [PubMed] [Google Scholar]
- 18.Riou C, Du Bruyn E, Ruzive S, et al. . Disease extent and anti-tubercular treatment response correlates with Mycobacterium tuberculosis-specific CD4 T-cell phenotype regardless of HIV-1 status. Clin Transl Immunol 2020; 9: e1176. doi: 10.1002/cti2.1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Yan LM, Wan L, et al. . Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 2020; 20: 656–657. doi: 10.1016/S1473-3099(20)30232-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Liu W, Zheng Y, et al. . A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect 2020; 22: 206–211. doi: 10.1016/j.micinf.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Chen C, Li Q, et al. . COVID-19 confirmed patients with negative antibodies results. BMC Infect Dis 2020; 20: 698. doi: 10.1186/s12879-020-05419-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juno JA, van Bockel D, Kent SJ, et al. . Cytotoxic CD4 T cells – friend or foe during viral infection? Front Immunol 2017; 8: 19. doi: 10.3389/fimmu.2017.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pachnio A, Ciaurriz M, Begum J, et al. . Cytomegalovirus infection leads to development of high frequencies of cytotoxic virus-specific CD4+ T cells targeted to vascular endothelium. PLoS Pathog 2016; 12: e1005832. doi: 10.1371/journal.ppat.1005832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathew D, Giles JR, Baxter AE, et al. . Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020; 369: eabc8511. doi: 10.1126/science.abc8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Wherry EJ. T cell responses in patients with COVID-19. Nat Rev Immunol 2020; 20: 529–536. doi: 10.1038/s41577-020-0402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grifoni A, Weiskopf D, Ramirez SI, et al. . Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020; 181: 1489–1501. doi: 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. . Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020; 183: 158–168. doi: 10.1016/j.cell.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bert N, Tan AT, Kunasegaran K, et al. . SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020; 584: 457–462. doi: 10.1038/s41586-020-2550-z [DOI] [PubMed] [Google Scholar]
- 29.Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. . Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020; 183: 996–1012. doi: 10.1016/j.cell.2020.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. . Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020; 181: 1036–1045. doi: 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sattler A, Angermair S, Stockmann H, et al. . SARS-CoV-2 specific T-cell responses and correlations with COVID-19 patient predisposition. J Clin Invest 2020; 130: 6477–6489. doi: 10.1172/JCI140965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schub D, Klemis V, Schneitler S, et al. . High levels of SARS-CoV-2-specific T cells with restricted functionality in severe courses of COVID-19. JCI Insight 2020; 5: e142167. doi: 10.1172/jci.insight.142167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng HY, Zhang M, Yang CX, et al. . Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol 2020; 17: 541–543. doi: 10.1038/s41423-020-0401-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appay V, Dunbar PR, Callan M, et al. . Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med 2002; 8: 379–385. doi: 10.1038/nm0402-379 [DOI] [PubMed] [Google Scholar]
- 35.Parry HM, Dowell AC, Zuo J, et al. . PD-1 is imprinted on cytomegalovirus-specific CD4+ T cells and attenuates Th1 cytokine production whilst maintaining cytotoxicity. PLoS Pathog 2021; 17: e1009349. doi: 10.1371/journal.ppat.1009349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasprowicz V, Schulze Zur Wiesch J, Kuntzen T, et al. . High level of PD-1 expression on hepatitis C virus (HCV)-specific CD8+ and CD4+ T cells during acute HCV infection, irrespective of clinical outcome. J Virol 2008; 82: 3154–3160. doi: 10.1128/JVI.02474-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jubel JM, Barbati ZR, Burger C, et al. . The role of PD-1 in acute and chronic infection. Front Immunol 2020; 11: 487. doi: 10.3389/fimmu.2020.00487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rha MS, Jeong HW, Ko JH, et al. . PD-1-expressing SARS-CoV-2-specific CD8+ T cells are not exhausted, but functional in patients with COVID-19. Immunity 2021; 54: 44–52. doi: 10.1016/j.immuni.2020.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murugesan K, Jagannathan P, Pham TD, et al. . Interferon-γ release assay for accurate detection of severe acute respiratory syndrome coronavirus 2 T-cell response. Clin Infect Dis 2021; 73: e3130–e3132. doi: 10.1093/cid/ciaa1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braudeau C, Salabert-Le Guen N, Chevreuil J, et al. . An easy and reliable whole blood freezing method for flow cytometry immuno-phenotyping and functional analyses. Cytometry B Clin Cytom 2021; 100: 652–665. doi: 10.1002/cyto.b.21994 [DOI] [PubMed] [Google Scholar]
- 41.Ogbe A, Kronsteiner B, Skelly DT, et al. . T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat Commun 2021; 12: 2055. doi: 10.1038/s41467-021-21856-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00285-2021.SUPPLEMENT (479.9KB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00285-2021.Shareable (334.4KB, pdf)