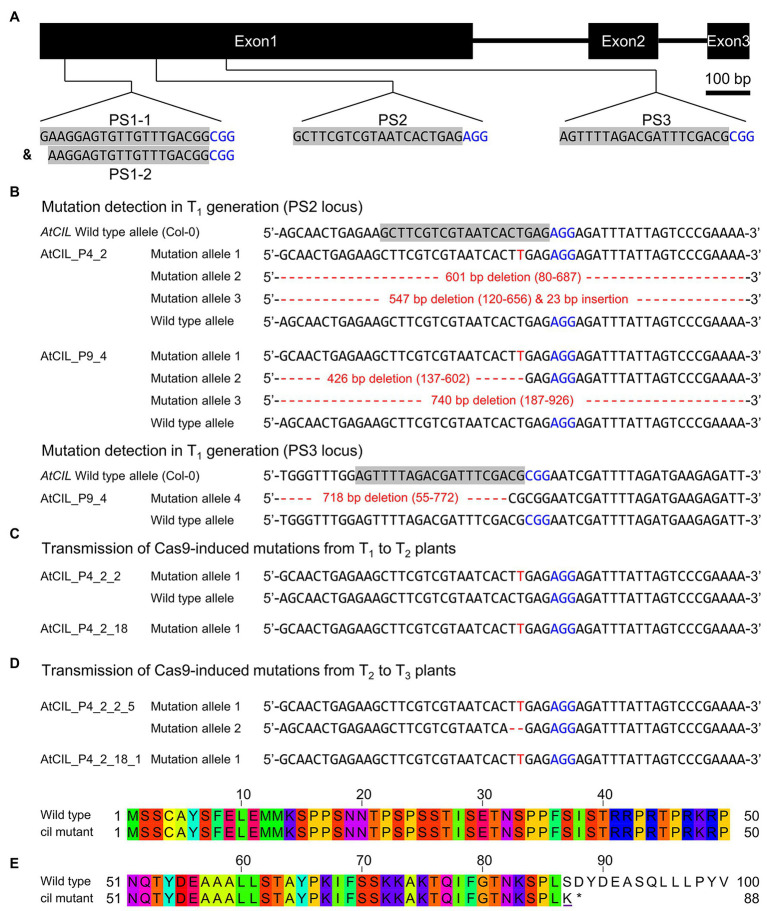
Figure 2.
Site-directed mutagenesis of the cil gene by CRISPR. (A) Structure of the cil gene and guide RNA selection. The protospacer sequences are indicated by gray background and the protospacer adjacent motifs are highlighted by blue color. (B) Mutation detection in T1 plants. Two chimeric plants, AtCIL_P4_2 and AtCIL_P9_4, carrying mutations at either or both PS2 and PS3 target loci. Large deletions/insertions are represented by red dashes and small insertions by red letters. Coordinates in parentheses indicate the region of deletion. The adenine in the start codon of the cil sequence is counted as +1. (C) Inheritance of mutations in the T2 generation. The heterozygous plant AtCIL_P4_2_2 and the homozygous plant AtCIL_P4_2_18 carry a one-nucleotide insertion 3 bp upstream of the protospacer adjacent motif of the PS2 locus. (D) Inheritance of mutations in the T3 generation. One biallelic mutant, AtCIL_P4_2_2_5, was identified. It carries a one-nucleotide insertion and a 2 bp deletion. The mutation of the T2 plant AtCIL_P4_2_18 is stably transmitted from the T2 to the T3 generation. (E) Alignment of the deduced N-terminal protein sequences of wild type and cil. The altered amino acid of the mutant is underlined; an asterisk indicates the position of the immature stop codon.