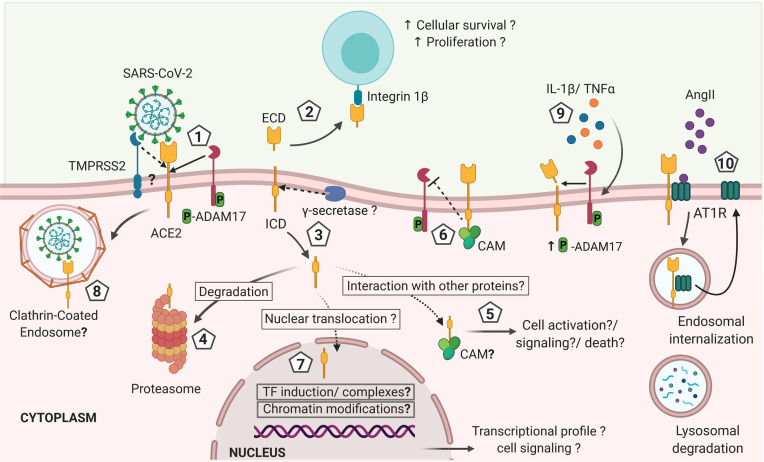
Figure 2.
Hypothetical model of ACE2 shedding by ADAM17 and γ-secretase after ACE2 interaction with SARS-CoV-2 and its effects on the immune response against the infection. 1). Initial interaction between SARS-CoV-2 spike protein and the transmembrane cellular receptor ACE2 may trigger proteolytic cleavage of ACE2 mediated by ADAM17, releasing the extracellular domain of this receptor (ectodomain). Other proteases such as TMPRSS2, HAT and Hepsin may also cleave ACE2 at a different catalytic site than ADAM17, thus potentially exhibiting differential outcomes. 2) Soluble ACE2 can interact with other cells through integrins acting like a cell adhesion molecule and triggering signals associated with cell proliferation and survival. 3) The cytoplasmic domain is likely released from the cellular membrane by γ-secretase, as described for other RIP proteins and it is either: 4) degraded at the proteasome, or 5) interacts with cytoplasmic proteins as it has been described for calmodulin (CAM) that can have effects on cellular activation, inflammation and cells death. 6) If CAM interacts with the transmembrane full-length ACE2, the shedding of ACE2 may be abrogated. 7) The intracellularly released ACE2 domain may also be translocated into the nuclei. At the nuclei, the endodomain may induce transcriptional expression of different genes by a still unknown mechanism that can include induction of transcription factors or formation of transcription factors-like complexes or chromatin modifications, influencing the cellular response and fate. In addition, the increased levels of IL-1β and TNFα induced during COVID-19 may increase ADAM17 phosphorylation and activity favoring a sustained shedding of ACE2. Thus, the shedding of ACE2 induced by SARS-CoV-2 infection is likely influencing the outcome of COVID-19 by mechanisms that are still unknown and that remain to be explored in order to design preventive and therapeutic approaches (created with BioRender.com).