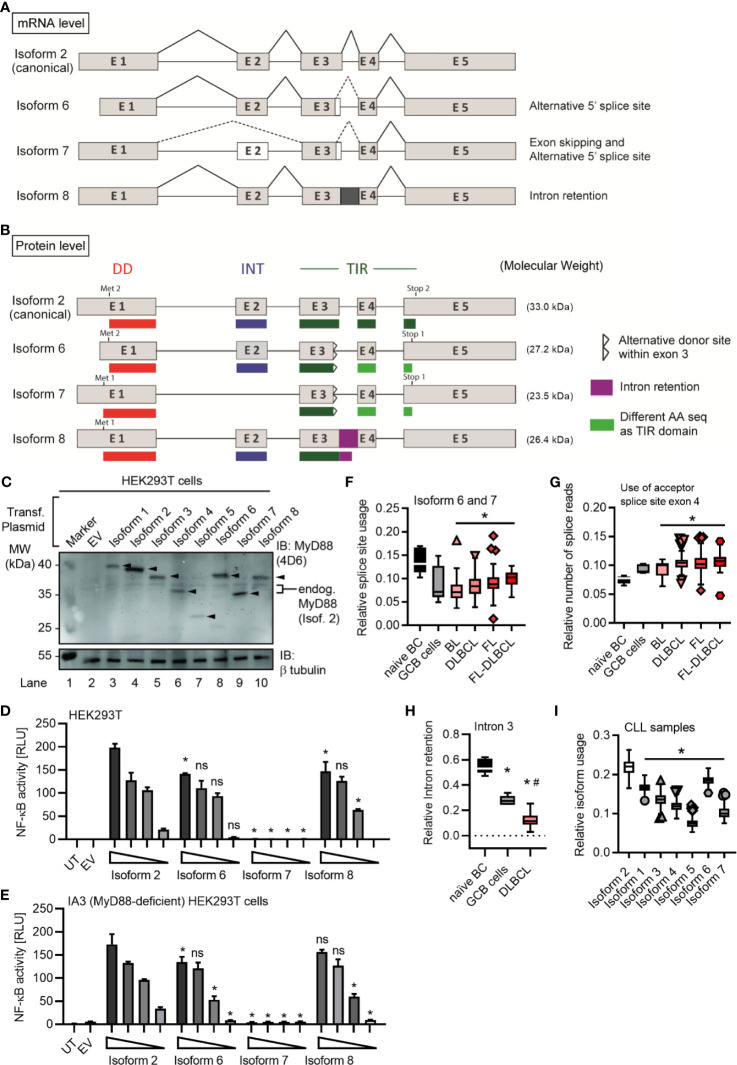
Figure 4.
MYD88 can give rise to three additional MyD88 isoforms. (A, B) Schematic representation of novel MYD88 isoforms on mRNA and protein level according to references in Table 1 and hypothetical sequence for isoform 8. (C–E) HEK293T cells were transfected with plasmids for different MYD88 isoforms and lysates analyzed for expression or pathway activation by immunoblot (C, n=2) or NF-κB dual luciferase assay (D, E n=3), respectively. (E) as in D but using MyD88-deficient I3A cells (n=3). (F–H) RNAseq analysis from untransformed B cells or lymphoma samples (n=as indicated in Figure 2E ). Intron retention presented as relative number of splice reads using the acceptor splice site of exon 4 (G) or coverage of intron 3 compared to mean of flanking exons 3 and 4 (H). (I) RNAseq analysis from CLL samples (n=289). In C–E one representative of ‘n’ technical replicates is shown, for D, E, as mean + SD from three repeats. F–I represent combined data (Tukey box and whiskers) from ‘n’ biological replicates (each dot represents one replicate). ns, non-significant; * or # = p<0.05 according to two-way ANOVA comparing to isoform 2 (D, E) or Wilcoxon Mann-Whitney (F-I) in comparison to naïve B cells (*, F–H) and to GCB cells (#, F–H) or isoform 2 (I).