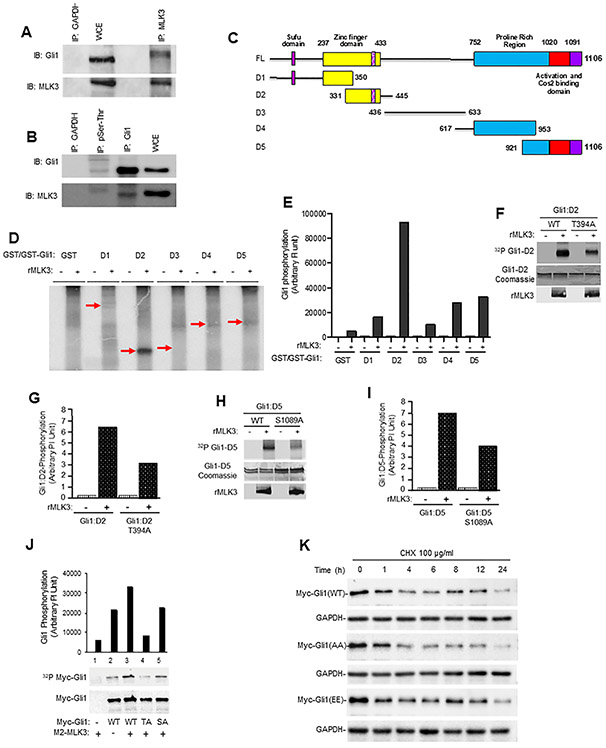
Figure 3. MLK3 associates with GLI1 and phosphorylates at Thr394 and Ser1089.
(A) The endogenous MLK3 protein from PANC-1 cell extracts was immunoprecipitated (I.P.) and blotted with an anti-GLI1 antibody. GAPDH antibody was used for control IP, and whole-cell extracts (WCE) were used for input. The membranes were blotted with GLI1 and MLK3 antibodies, respectively. (B) The cell extract from PANC-1 cells was immunoprecipitated either with pSer/pThr or anti-GLI1, or anti-GAPDH (control) and cell extract (30 μg) was used as an input, and membranes were blotted with either GLI1 or MLK3 antibodies. Phosphorylation-dependent interaction between MLK3 and GLI1 was performed using the pSer/pThr antibody. (C) Schema detailing the five GLI1 deletion mutants (i.e., D1-D5) and Full length (FL). (D-E) An in vitro kinase assay was performed using bacterially expressed GLI1 deletion mutant (D1-D5) proteins and recombinant MLK3 enzyme, values presented as an arbitrary unit. (F-I) Bacterially expressed wild type GLI1 (WT), GLI1-T394A, or GLI1-S1089A with and without recombinant MLK3 (rMLK3) enzyme were used in in vitro kinase assays. Phosphorylation of these GLI1 fragments was determined by Phosphoimager and presented as an arbitrary unit (g and i). (J) The confirmation of GLI1 -T394 and -S1089 as the in vivo targets of MLK3 was determined in HEK-293 cells after cell labeling with P32-orthophosphate. (K) The stability of indicated GLI1 protein variants (GLI1 -WT, -AA and -EE) was determined by cycloheximide chase assay, followed by western blotting with an anti-Myc antibody for transfected Myc-tagged GLI1 proteins.