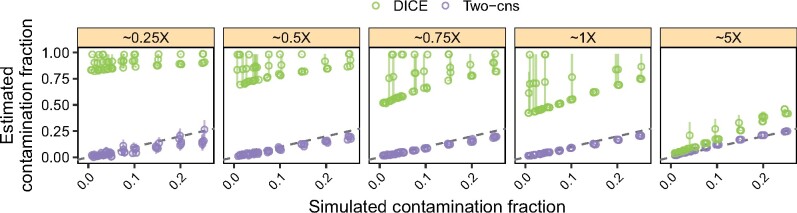
Fig. 2.
Simulation results comparing our method to DICE. We simulated data as described in Section 3.5 and estimated contamination across five replicates using our method (purple) and DICE (green). We ‘contaminated’ the Anzick1 ancient Native American genome with a French individual at increasing contamination fractions while controlling for the DoC. Vertical bars correspond to 95% confidence intervals for the Two-consensus method and to 95% credible intervals for DICE. The dashed line indicates the expected values. Note that the simulated DoC corresponds to the autosomal DoC for DICE and the X-chromosome DoC for our method (i.e. autosomes and X-chromosomes were simulated independently)