Figure 1:
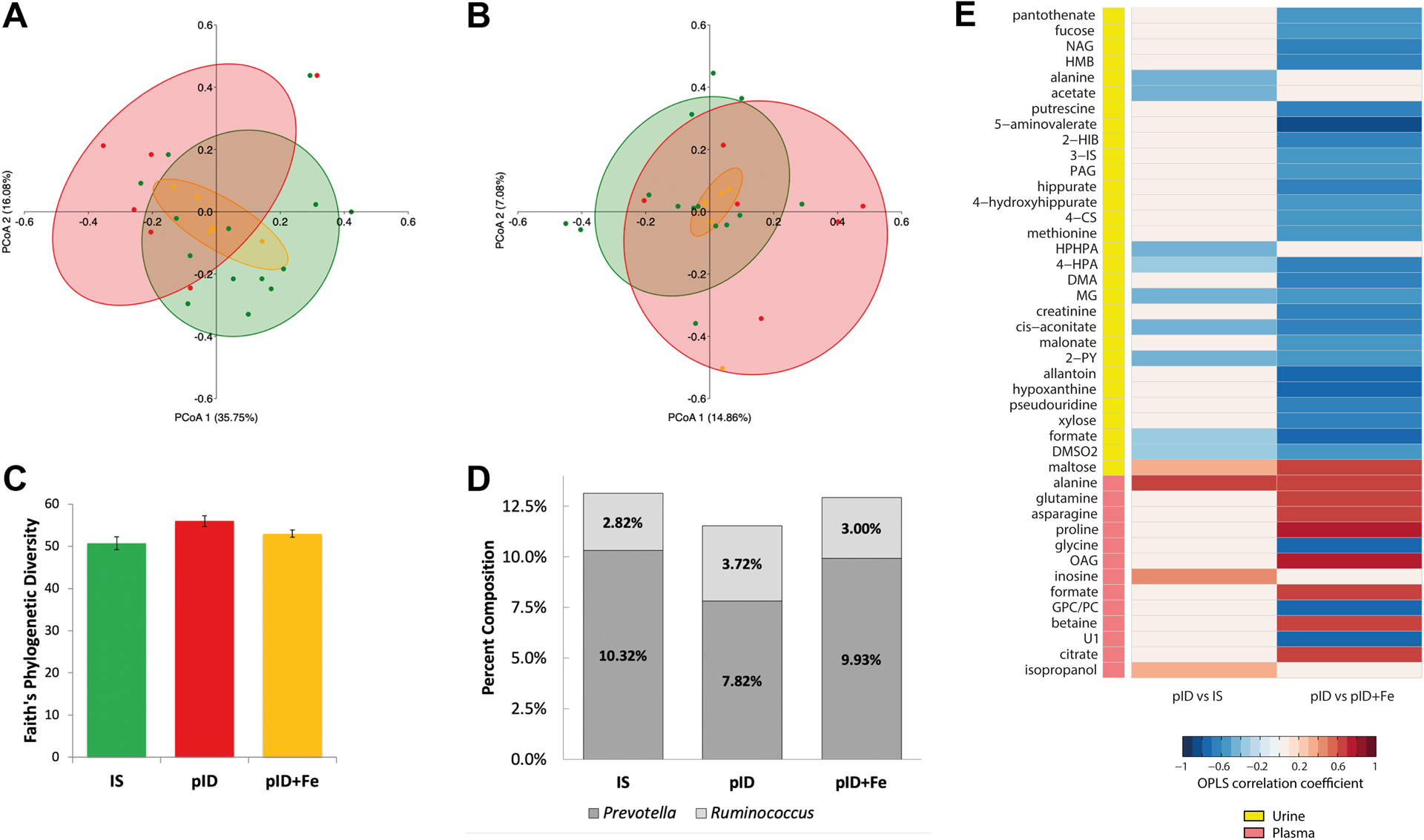
Variation in the community structure of the fecal microbiota measured by 16S rRNA gene sequencing and the urinary and plasma metabolic disruptions associated with gradual recovery from iron deficiency (pID) when compared to continuously iron sufficient (IS) and iron-supplemented monkeys (pID+Fe). (A) Weighted UniFrac analyses revealed significant albeit modest differences in the community structure between young monkeys that had recovered gradually from iron deficiency (pID, red) and those that remained iron sufficient (green, IS) or ones that received iron supplements when anemic (pID+Fe, yellow). Significance testing was performed by regressing PCoA axes 1 and 2 on group membership using planned orthogonal contrasts. (B) Differences were not evident with Unweighted UniFrac analyses indicating variation in community structure was attributable to the abundances, not the presence or absence of taxa. Percentages indicate variance in beta diversity variance accounted for by each PCoA axis. (C) Significant differences in the richness of the community structure were not apparent. (D) Prevotella and Ruminococcus were the two most abundant genera found to differ across the three conditions. Prevotella was highest in IS monkeys, and lowest in the pID animals; conversely, Ruminococus was lowest in IS and highest in the pID group. (E) Colors indicate correlation coefficients extracted from the OPLS-DA models comparing pID and IS urine (Q2Y = 0.10; p = 0.045), and plasma profiles (Q2Y = 0.29; p = 0.029) and the pID and pID+Fe urine (Q2Y = 0.21; p = 0.039) and plasma profiles (Q2Y = 0.45; p = 0.001). Red indicates metabolites that more abundant in the pID samples and blue indicates metabolites less abundant in the pID samples compared to the 2 other groups. The key indicates whether the metabolic change occurred in the urine (yellow) or plasma (pink).
