Figure 1.
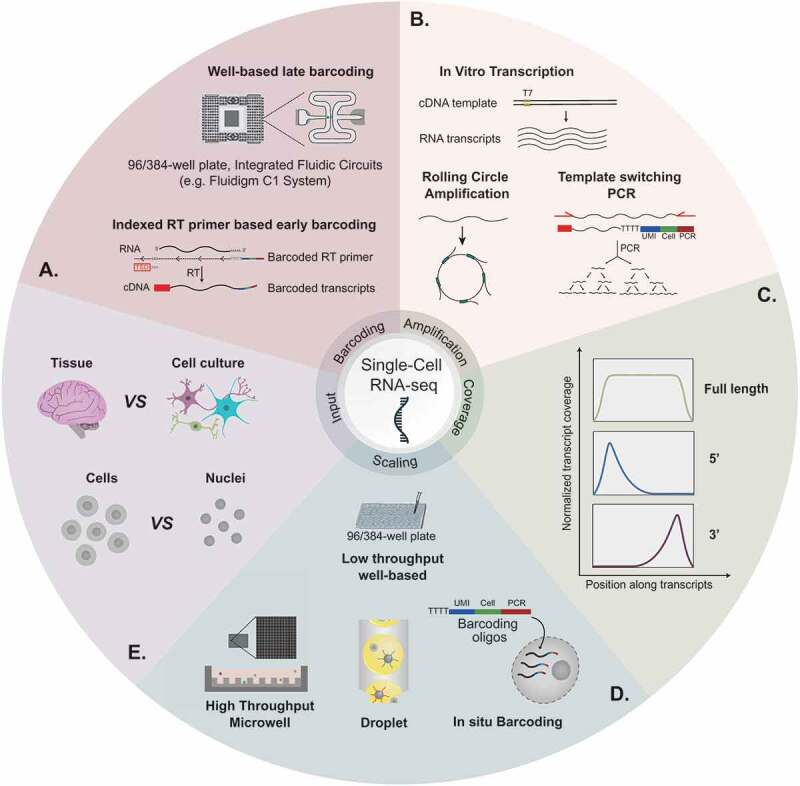
A recipe book for scRNA-seq: A) Cellular barcoding can be achieved by PCR or tagmentation-based late indexing of independent cDNA libraries prepared in separated chambers, or via early introduction of cell barcodes during the reverse transcription reaction. B) Amplification of low abundance cellular material can be achieved by in vitro transcription, PCR following a template-switching reverse transcription reaction, or rolling circle amplification. C) The choice of scRNA-seq workflow will determine the distinct coverage enrichment profiles observed along captured RNA transcripts. D) Independent cDNA libraries prepared in separated chambers can lead to full transcript coverage at reduced scale. Increased scaling can be achieved by increasing the number of ‘chambers’ from which libraries are prepared, and providing cellular indexes during reverse transcription such that early pooling of cellular transcriptomes can be achieved. The former can be achieved using massively parallel microwells, using microfluidic systems to rapidly generate thousands of oil droplet-based reaction chambers that encapsulate single cells, or employing the cells themselves as the reaction chambers for in situ library preparations. E) Starting material can be heterogeneous tissue or cell culture preparations. The characteristics of the input will determine whether whole cells or purified nuclei are to be profiled
