Figure 1. C. albicans sexual regulators control commensal fitness.
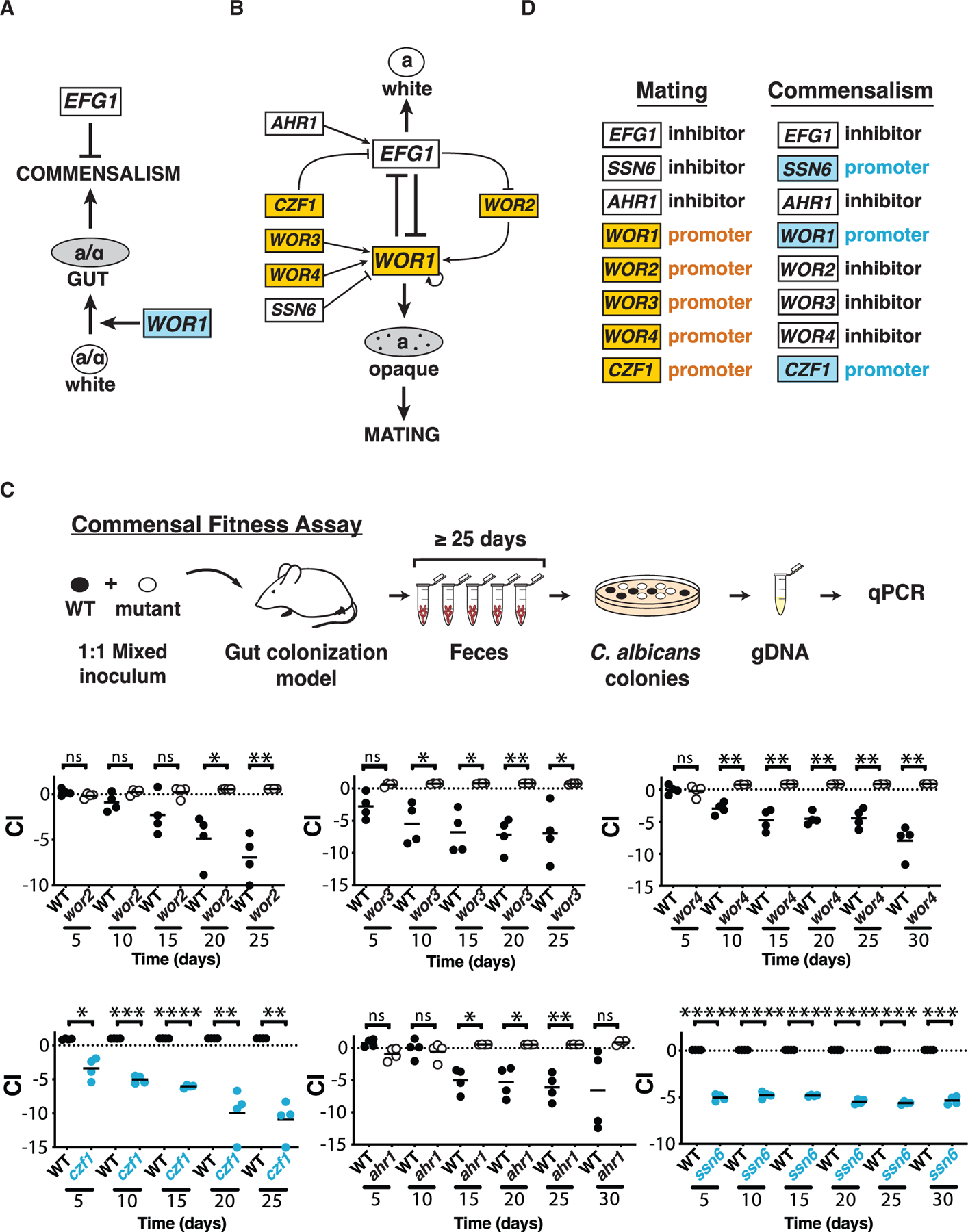
(A) Regulation of gut commensalism. Efg1 inhibits (bar) and Wor1 promotes (arrow) fungal commensalism (Pande et al., 2013). Wor1 promotes the white-to-gut developmental switch that confers enhanced fitness in the mammalian digestive tract.
(B) Regulation of mating. Efg1 is the central inhibitor and Wor1 the central activator of mating and the white-to-opaque switch. Six additional transcription factors interact with Efg1 and Wor1 via the depicted regulatory circuit (Hernday et al., 2013, 2016, 2010; Huang et al., 2006; Lohse et al., 2010; Tuch et al., 2010; Wang et al.; Zordan et al., 2006, 2007).
(C) WOR2, WOR3, WOR3, CZF1, AHR1, and SSN6 regulate the commensal fitness of C. albicans. Each of the indicated null mutants was tested in 1:1 competitions with WT in a mouse model of stable gut colonization. Mutants with loss-of-fitness phenotypes are shown in blue, and those with gain-of-fitness in white (with black outline). Statistical comparisons were performed using the paired Student’s t test; ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Results for independent isolates of these mutants are presented in Figure S2.
(D) Comparison of the roles of eight fungal transcription factors in mating versus commensalism.
