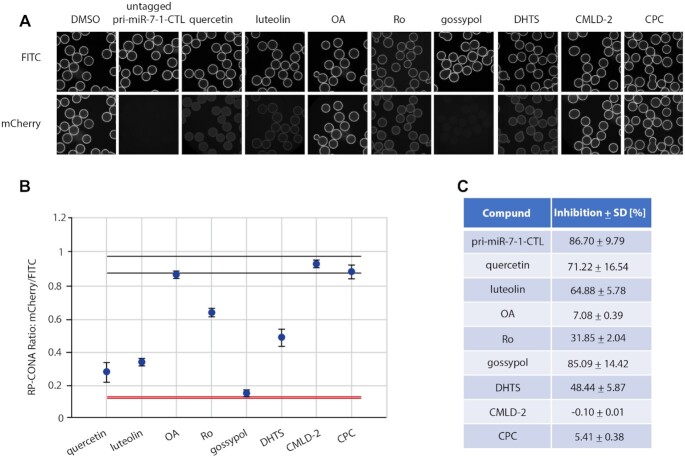
Figure 6.
RP-CONA identified pri-miR-7–1/HuR inhibitors from a focused library. (A) Cell lysates containing mCherry-HuR were treated with DMSO, 50 μM of untagged pri-miR-7–1-CTL or 100 μM of compounds before pulldown. Beads images taken in ImageXpress. (B) Relative mCherry/FITC ring intensity mean and SD between the beads in each well after compound treatment are shown. DMSO (RP-CONA ratio: 0.932 ± 0.016, CV: 1.75%, n = 5) served as a negative control while 50 μM of untagged pri-miR-7–1-CTL (RP-CONA ratio: 0.124 ± 0.001, CV: 0.96%, n = 5) served as a positive control. Z’ = 0.93. Black lines: DMSO mean ± 3 × SD between five repeated wells. Red lines: untagged pri-miR-7–1-CTL mean ± 3 × SD between five repeated wells. At least 400 beads were included in each control analysis. (C). Inhibition of RP-CONA signals compared to DMSO. The percentage inhibition of compounds relative to DMSO mean are shown. The SD of relative inhibitions between the beads in each well after compound treatment are shown.