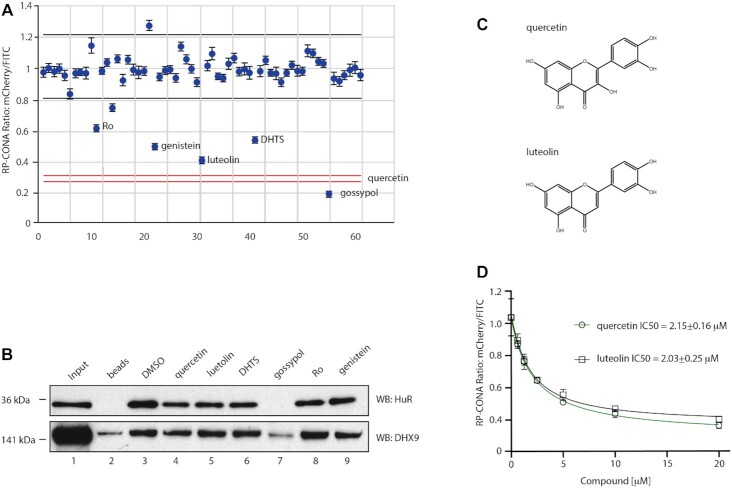
Figure 7.
Identification and validation of pri-miR-7–1/HuR inhibitors from an enlarged library. (A) A small-scale prototype RP-CONA screen to test the disruptive effects of 61 compounds (54 from an in-house library varied from 1 to 100 μM, and seven from the previous identified HuR or MSI2 inhibitors at 100 μM) on pri-miR-7–1/HuR. Relative mCherry/FITC ring intensity mean and SD between the beads in each well after compound treatment are shown. DMSO (RP-CONA ratio: 1.009 ± 0.067, CV: 6.70%, n = 6) served as a negative control while 100 μM of quercetin (RP-CONA ratio: 0.288 ± 0.006, CV: 2.18%, n = 6) served as a positive control. Z’ = 0.69. Black lines: DMSO mean ± 3 × SD between six repeated wells. Red lines: quercetin mean ± 3 × SD between six repeated wells. At least 500 beads were included in each control analysis. Compounds generating >40% (six times CV of negative controls) inhibition are annotated. (B) Disruption of endogenous HuR pulled down by pri-miR-7–1-CTL. Pri-miR-7–1-CTL was covalently linked to agarose beads. HeLa extracts were treated with DMSO or 100 μM of each compound prior to pull-down. HuR and DHX9 in the pull-down components were detected by western blot. 1: Input. Unbound proteins in the lysates after pulldown. 2: Proteins pulled down by beads without RNA. 3–9: Proteins pulled down by pri-miR-7–1-CTL-beads after compound treatment. (C) Chemical structures of quercetin and luteolin. (D) Quercetin and luteolin disrupted pri-miR-7/HuR in a dose-dependent manner in RP-CONA. Increased concentrations of quercetin and luteolin were tested in RP-CONA. Relative mCherry/FITC ring intensity mean and SD between triplicated wells after compound treatment are shown. The RP-CONA signals were curve fitted by non-linear regression-four parameter [inhibitor] versus response, and IC50s were determined with the 4-parameter equation in GraFit v7.0.3 (Erithacus Software Limited) (40). The IC50s of quercetin and luteolin were 2.15 ± 0.16 μM and 2.03 ± 0.25 μM, respectively.