Figure 6.
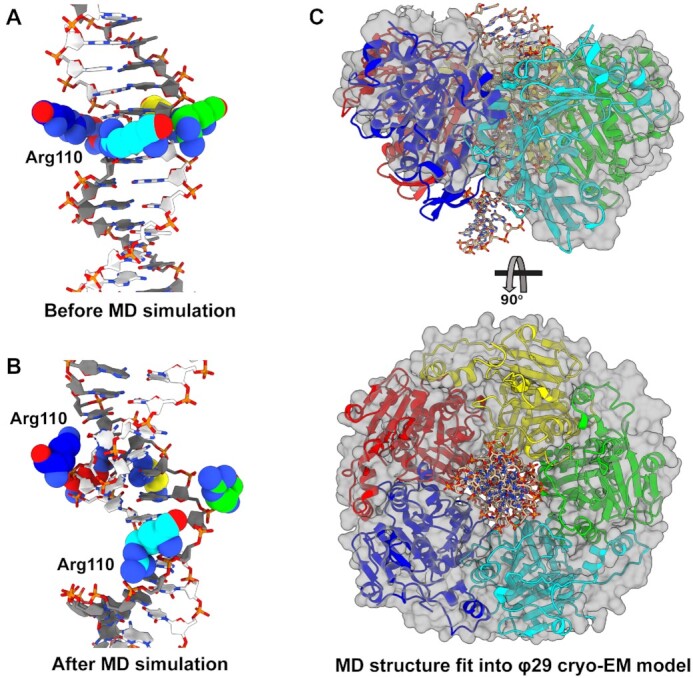
Predicted helicity of asccφ28 ATPase domains. (A) Initial set up of the asccφ28 NTD ring with DNA in the pore, prior to MD simulation. Initially, all five Arg110s are in a planar ring. (B) The predicted configuration after MD simulation. The five Arg110s adopt a helical pitch complementary to DNA, primarily tracking one strand of DNA. The Arg110s approach the phosphates differently: the lower three subunits (cyan, green, yellow) track ‘above’ the dark gray strand every two base pairs. The remaining two subunits (red, blue) fit into the minor groove and contact the dark gray strand from ‘below’, while also interacting with nucleobases. (C) The MD simulated asccφ28 NTD ring was fit into the cryo-EM reconstruction of φ29 stalled during packaging. The asccφ28 ring is shown as Richardson diagrams, DNA as sticks, and the model of φ29 NTDs built into the reconstruction as a translucent surface.
