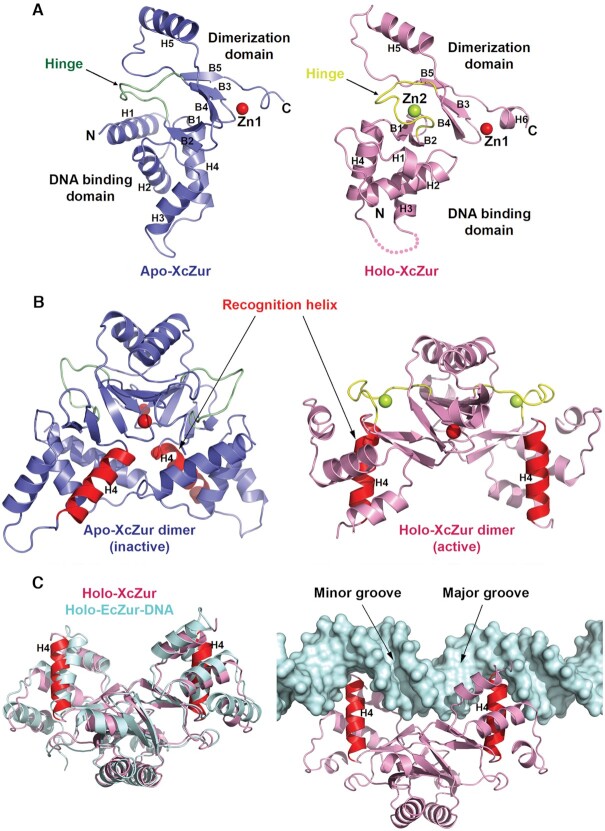
Figure 3.
Apo- and holo-XcZur structures reveal distinct competencies for DNA binding. (A) Cartoon representation of the apo- and holo-XcZur monomers. The apo-XcZur structure is shown in blue, with the hinge region highlighted in green (left panel). The holo-XcZur structure is shown in pink, with the hinge highlighted in yellow (right panel). Zn1 and Zn2 are shown in red and limon spheres, respectively. Secondary structural elements are labeled. (B) Cartoon representation of the apo- (left panel) and holo-XcZur (right panel) dimers. Recognition helix (H4) is highlighted in red. (C) Structural comparison of the holo-XcZur dimer with the holo-EcZur structure from the EcZur-DNA complex. Left panel shows a superimposition of holo XcZur (pink) with holo EcZur (cyan). Right panel displays a docking of holo XcZur into the EcZur-box (cyan surface) in the same orientation of EcZur. Minor and major grooves of the DNA are indicated.